zgc:110425
ZFIN 
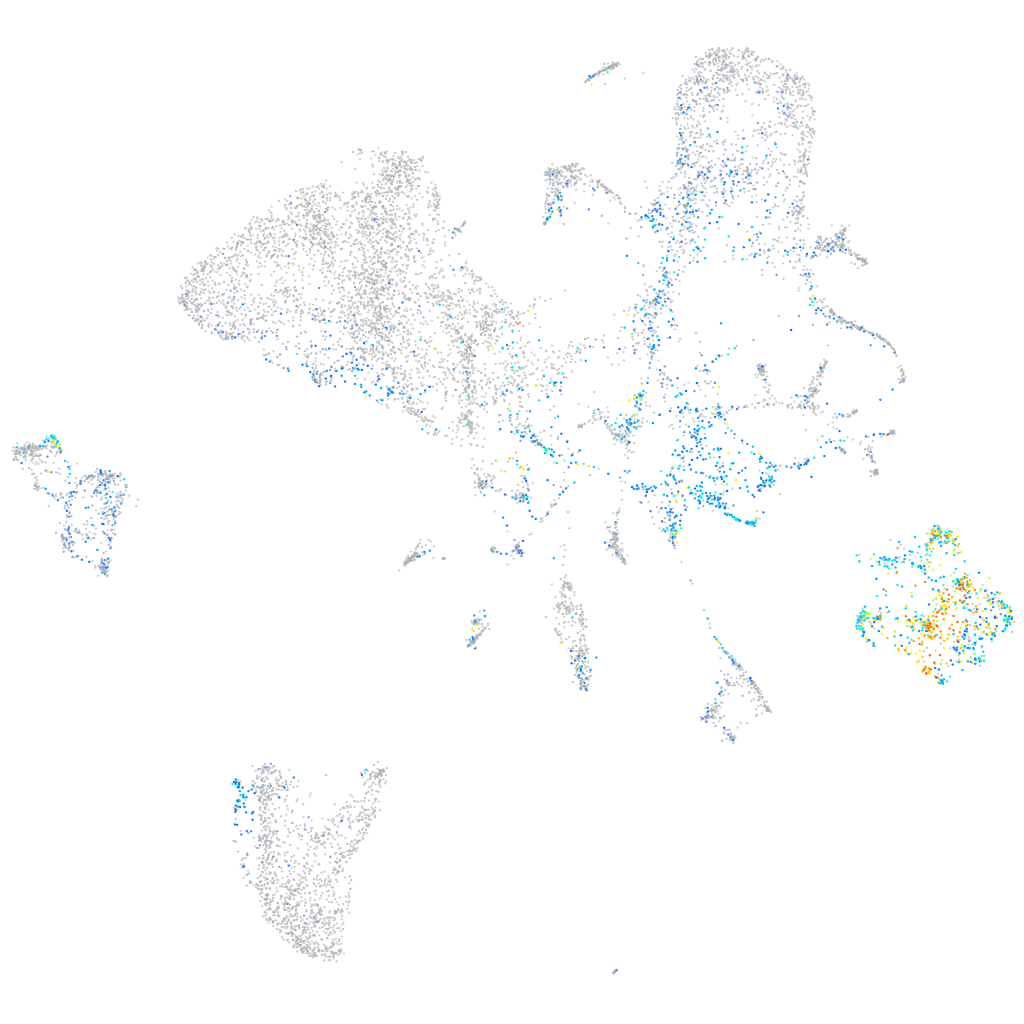
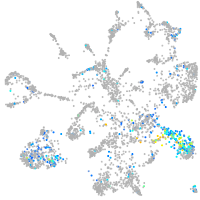
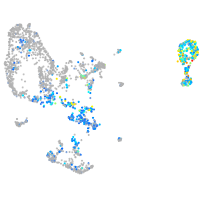
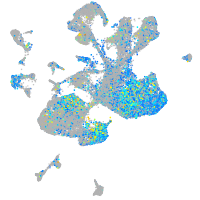
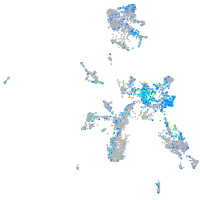
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-108k21.10 | 0.652 | rpl37 | -0.482 |
| zgc:110216 | 0.651 | rps10 | -0.464 |
| si:ch211-113a14.18 | 0.641 | zgc:114188 | -0.431 |
| mki67 | 0.626 | rps17 | -0.400 |
| akap12b | 0.624 | gamt | -0.374 |
| hspb1 | 0.616 | nme2b.1 | -0.373 |
| marcksb | 0.606 | gapdh | -0.372 |
| si:ch211-152c2.3 | 0.601 | ahcy | -0.365 |
| seta | 0.596 | eno3 | -0.347 |
| tpx2 | 0.591 | zgc:92744 | -0.337 |
| lig1 | 0.590 | gatm | -0.330 |
| hnrnpa1b | 0.589 | sod1 | -0.328 |
| zgc:153405 | 0.588 | gpx4a | -0.316 |
| anp32a | 0.585 | atp5if1b | -0.315 |
| brd3a | 0.584 | suclg1 | -0.314 |
| nucks1a | 0.584 | pklr | -0.311 |
| dek | 0.584 | eef1da | -0.304 |
| hmga1a | 0.582 | bhmt | -0.304 |
| chaf1a | 0.582 | nupr1b | -0.301 |
| ppig | 0.580 | apoa1b | -0.301 |
| hmgb2a | 0.580 | glud1b | -0.295 |
| cx43.4 | 0.578 | apoa4b.1 | -0.294 |
| syncrip | 0.578 | tpi1b | -0.293 |
| anp32b | 0.577 | fbp1b | -0.292 |
| hnrnpub | 0.574 | mat1a | -0.292 |
| ilf3b | 0.572 | prdx2 | -0.290 |
| smc1al | 0.569 | zgc:158463 | -0.285 |
| acin1a | 0.567 | apoa2 | -0.282 |
| marcksl1b | 0.566 | pnp4b | -0.281 |
| si:ch211-113a14.24 | 0.561 | aldob | -0.278 |
| si:ch73-281n10.2 | 0.557 | atp5l | -0.278 |
| cbx3a | 0.552 | dap | -0.278 |
| NC-002333.4 | 0.551 | cx32.3 | -0.278 |
| hdgfl2 | 0.548 | mt-atp6 | -0.278 |
| hnrnpa1a | 0.547 | ckba | -0.278 |