zgc:100906 [Source:ZFIN;Acc:ZDB-GENE-040801-20]
ZFIN 
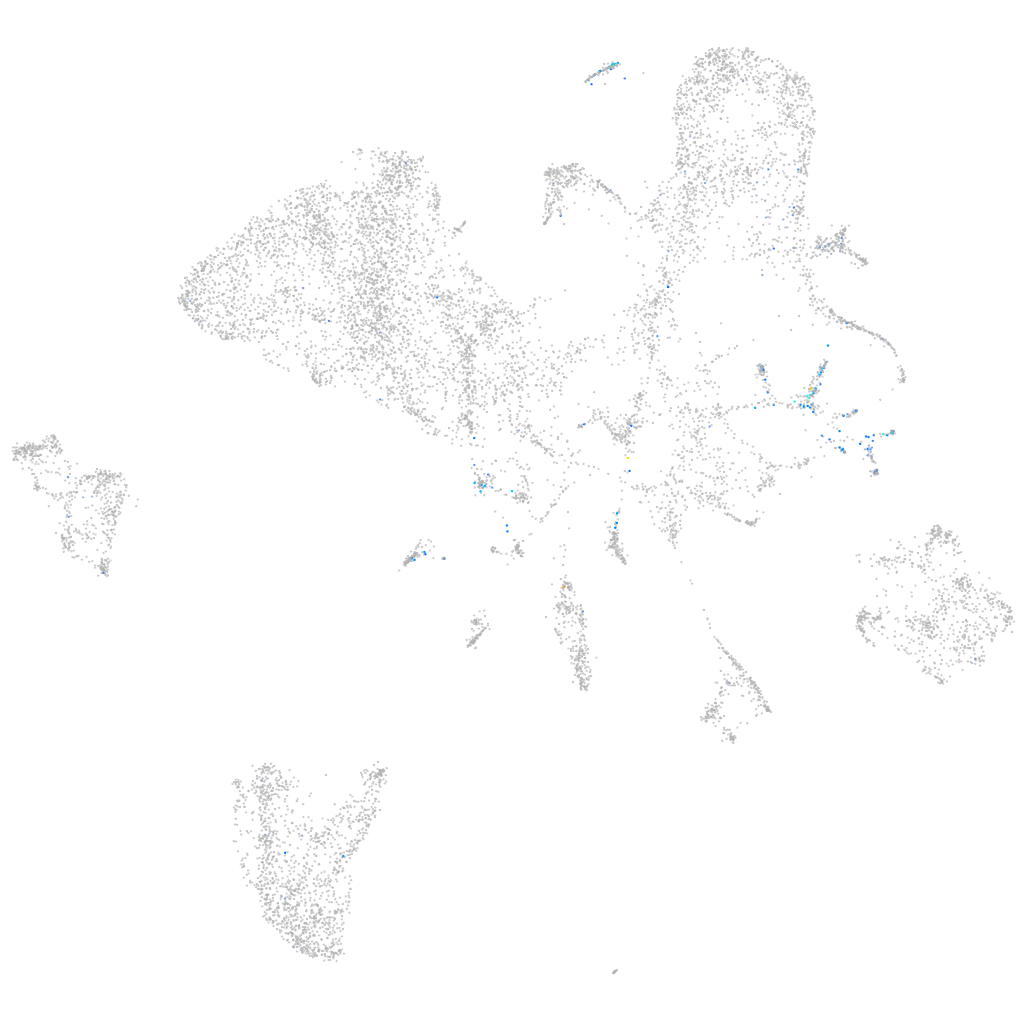
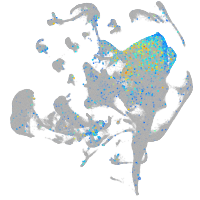
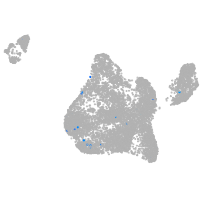
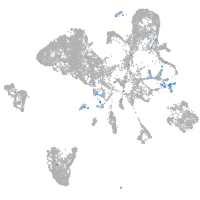
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.316 | ahcy | -0.093 |
| pcsk1nl | 0.310 | gamt | -0.092 |
| zgc:101731 | 0.279 | gapdh | -0.087 |
| egr4 | 0.264 | hdlbpa | -0.078 |
| neurod1 | 0.254 | aldh7a1 | -0.078 |
| syt1a | 0.247 | fabp3 | -0.077 |
| chga | 0.245 | gatm | -0.076 |
| si:dkey-153k10.9 | 0.243 | mat1a | -0.076 |
| mir7a-1 | 0.242 | rpl4 | -0.075 |
| insm1b | 0.239 | gstr | -0.075 |
| fev | 0.237 | si:dkey-16p21.8 | -0.073 |
| c2cd4a | 0.237 | bhmt | -0.072 |
| rprmb | 0.235 | agxtb | -0.071 |
| si:ch73-160i9.2 | 0.234 | aqp12 | -0.070 |
| hepacam2 | 0.230 | aldob | -0.070 |
| pcsk1 | 0.230 | agxta | -0.069 |
| id4 | 0.229 | mgst1.2 | -0.069 |
| rnasekb | 0.227 | apoa4b.1 | -0.068 |
| nmbb | 0.226 | aldh6a1 | -0.068 |
| scg2b | 0.222 | nupr1b | -0.067 |
| tspan7b | 0.222 | fbp1b | -0.067 |
| myt1b | 0.221 | si:dkey-151g10.6 | -0.067 |
| mir375-2 | 0.219 | cx32.3 | -0.067 |
| insm1a | 0.217 | apoc2 | -0.067 |
| nucb2b | 0.217 | shmt1 | -0.067 |
| atp6v0cb | 0.214 | apoa1b | -0.066 |
| vamp2 | 0.214 | gstt1a | -0.066 |
| pcsk2 | 0.213 | scp2a | -0.064 |
| vat1 | 0.213 | pnp4b | -0.064 |
| scgn | 0.213 | apoc1 | -0.063 |
| hsbp1a | 0.212 | gcshb | -0.063 |
| mapk15 | 0.208 | wu:fj16a03 | -0.063 |
| nsfa | 0.207 | rpl6 | -0.063 |
| kcnj11 | 0.207 | ttc36 | -0.063 |
| gdi1 | 0.207 | serpina1l | -0.062 |