zinc finger DHHC-type palmitoyltransferase 20a
ZFIN 
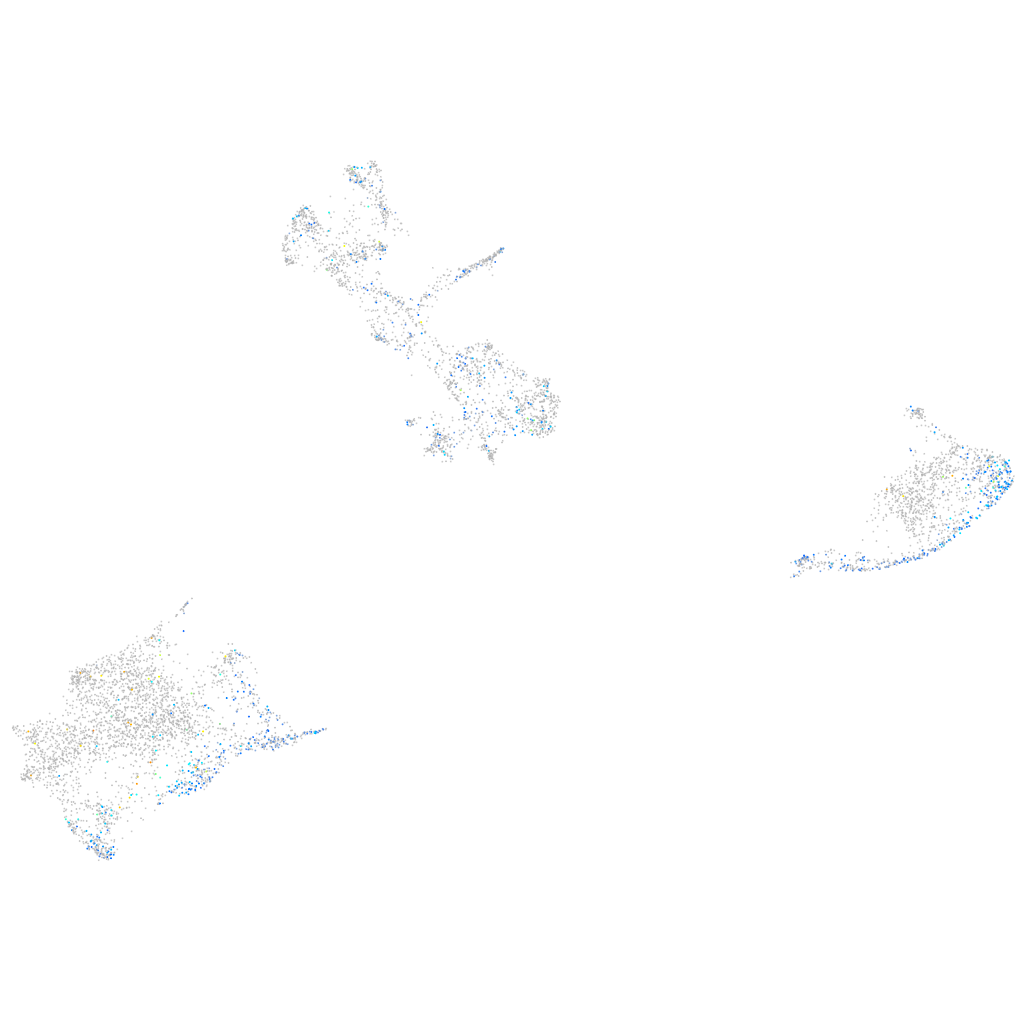
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.180 | pvalb2 | -0.061 |
| CR847566.1 | 0.157 | si:dkey-151g10.6 | -0.058 |
| CR383676.1 | 0.154 | mylpfa | -0.053 |
| trpm1b | 0.153 | pvalb1 | -0.052 |
| pcdh10a | 0.144 | uraha | -0.050 |
| pim1 | 0.142 | mylz3 | -0.047 |
| myo5aa | 0.142 | tmsb4x | -0.046 |
| ctsla | 0.142 | myhz1.1 | -0.045 |
| cdh1 | 0.141 | hbbe1.3 | -0.044 |
| XLOC-020079 | 0.141 | krt4 | -0.044 |
| opn5 | 0.141 | ttn.2 | -0.044 |
| sik1 | 0.140 | rpl10 | -0.043 |
| slc7a5 | 0.139 | tnnc2 | -0.042 |
| si:zfos-943e10.1 | 0.137 | rps12 | -0.042 |
| aatkb | 0.137 | ptmaa | -0.042 |
| kita | 0.137 | krt5 | -0.039 |
| bace2 | 0.136 | rpl7 | -0.039 |
| klf6a | 0.134 | rps23 | -0.037 |
| pcdh9 | 0.131 | actc1b | -0.037 |
| inka1b | 0.129 | tpma | -0.037 |
| dglucy | 0.128 | hbae3 | -0.037 |
| rasd1 | 0.127 | tmsb | -0.037 |
| mchr2 | 0.126 | rps19 | -0.035 |
| kcnj13 | 0.123 | paics | -0.035 |
| gadd45bb | 0.123 | fabp7a | -0.035 |
| lamp1a | 0.123 | atp2a1 | -0.035 |
| pknox1.2 | 0.122 | hbae1.1 | -0.034 |
| sdc4 | 0.122 | acta1b | -0.034 |
| atp6v0a2b | 0.122 | rpl19 | -0.034 |
| tfap2e | 0.122 | rplp1 | -0.034 |
| ddit3 | 0.121 | defbl1 | -0.034 |
| zdhhc2 | 0.121 | cyt1 | -0.033 |
| pah | 0.120 | hbbe1.2 | -0.033 |
| si:ch211-195b13.1 | 0.119 | apoda.1 | -0.033 |
| slc24a4a | 0.118 | rps20 | -0.033 |