"zinc finger, C4H2 domain containing"
ZFIN 
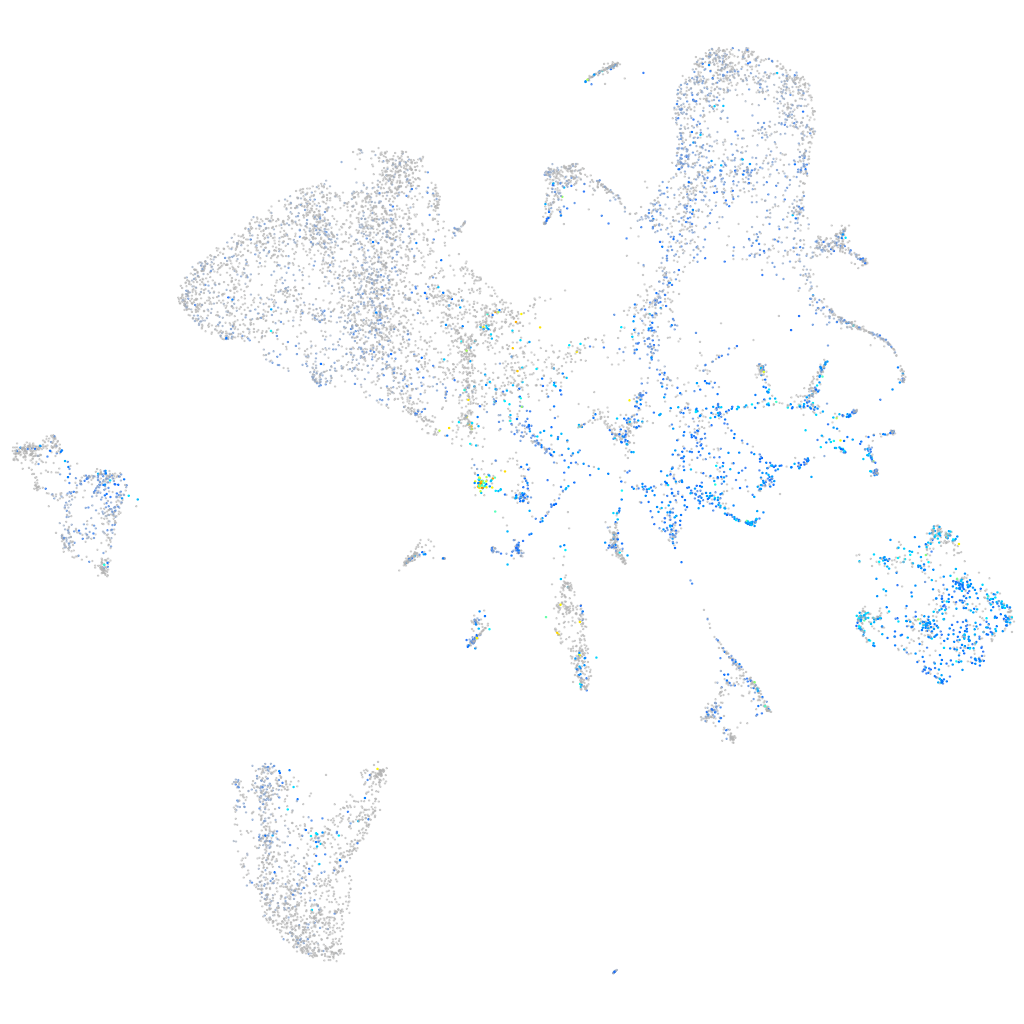
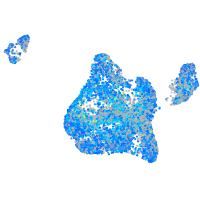
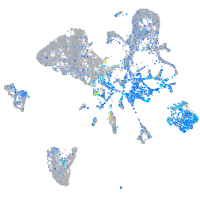
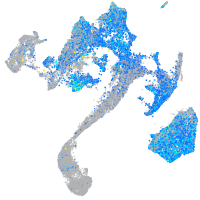
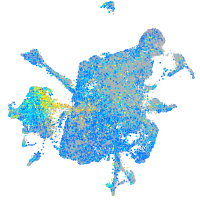
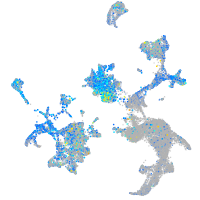
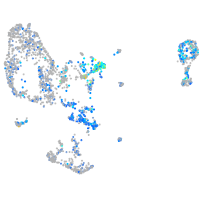
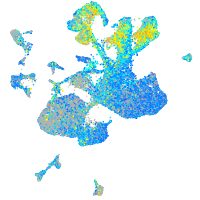
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb3a | 0.430 | gapdh | -0.318 |
| hnrnpaba | 0.429 | gamt | -0.310 |
| marcksl1b | 0.427 | ahcy | -0.305 |
| hmgn6 | 0.426 | gatm | -0.289 |
| khdrbs1a | 0.423 | nupr1b | -0.273 |
| h3f3d | 0.422 | mat1a | -0.264 |
| si:ch211-222l21.1 | 0.421 | eno3 | -0.263 |
| hdac1 | 0.413 | pnp4b | -0.261 |
| marcksb | 0.410 | agxtb | -0.261 |
| hnrnpa0a | 0.407 | fbp1b | -0.260 |
| si:ch73-1a9.3 | 0.405 | aldob | -0.260 |
| hmgb1b | 0.402 | bhmt | -0.255 |
| cirbpb | 0.398 | gpx4a | -0.254 |
| h2afvb | 0.391 | sod1 | -0.253 |
| hnrnpabb | 0.391 | aldh6a1 | -0.246 |
| hmga1a | 0.391 | apoa1b | -0.245 |
| nucks1a | 0.389 | glud1b | -0.244 |
| hnrnpa0b | 0.389 | apoa4b.1 | -0.244 |
| hmgn7 | 0.382 | scp2a | -0.243 |
| ptmab | 0.376 | ckba | -0.242 |
| sumo3a | 0.373 | abat | -0.241 |
| syncrip | 0.373 | suclg1 | -0.240 |
| si:ch211-288g17.3 | 0.373 | cx32.3 | -0.239 |
| hp1bp3 | 0.371 | eef1da | -0.236 |
| smarce1 | 0.369 | dap | -0.233 |
| cirbpa | 0.364 | zgc:92744 | -0.231 |
| si:ch73-281n10.2 | 0.362 | fabp10a | -0.231 |
| ilf3b | 0.360 | aldh7a1 | -0.231 |
| seta | 0.359 | g6pca.2 | -0.230 |
| hmgb2b | 0.354 | gnmt | -0.230 |
| ilf2 | 0.354 | grhprb | -0.229 |
| nono | 0.354 | apoa2 | -0.228 |
| cbx3a | 0.353 | rbp4 | -0.228 |
| cx43.4 | 0.350 | apom | -0.227 |
| anp32a | 0.350 | ces2 | -0.225 |