zinc finger and BTB domain containing 22a
ZFIN 
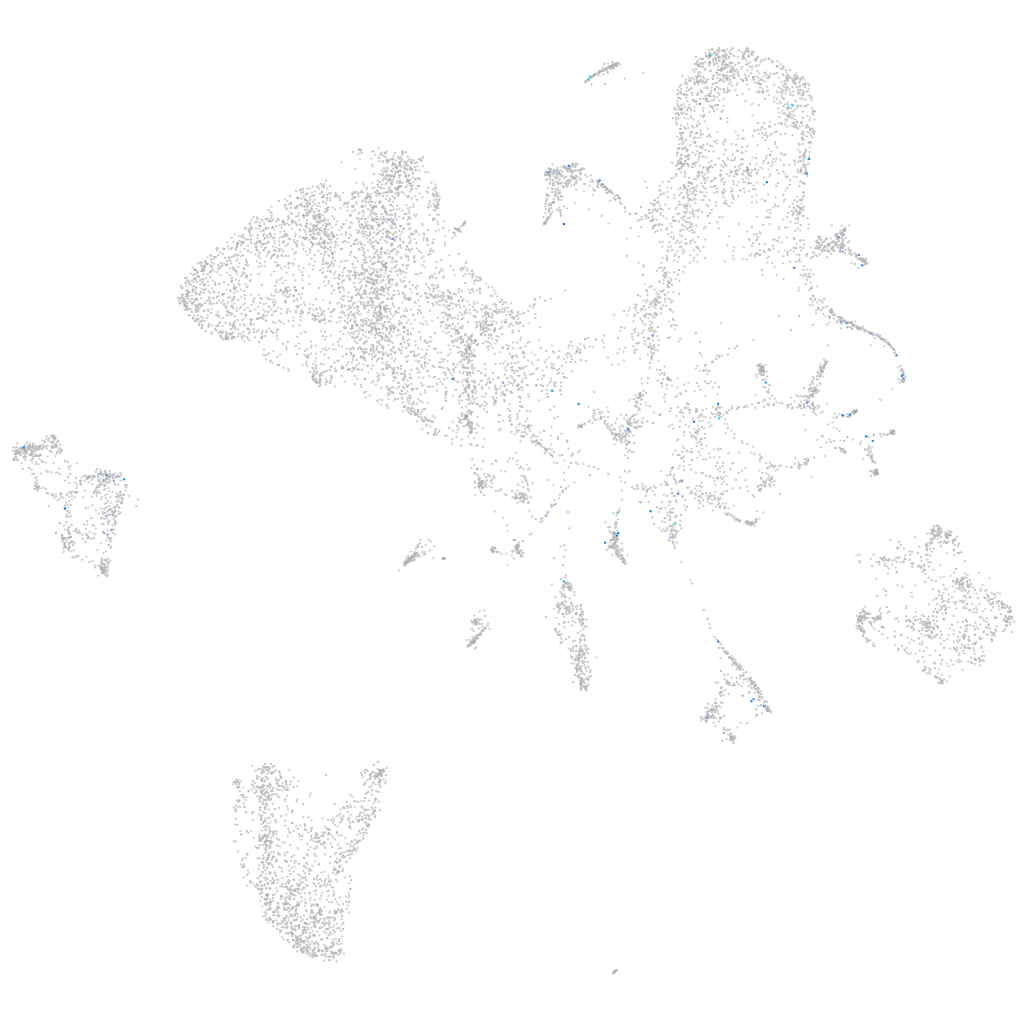
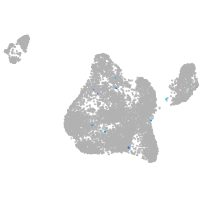
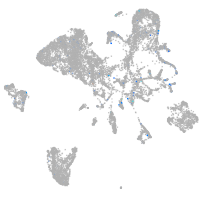
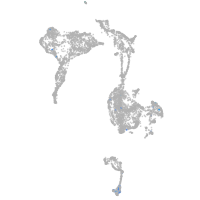
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108179280 | 0.358 | gamt | -0.055 |
| tmtops2b | 0.249 | aqp12 | -0.052 |
| BX248128.2 | 0.234 | ahcy | -0.052 |
| LOC556170 | 0.218 | bhmt | -0.049 |
| guca1c | 0.212 | apoc1 | -0.049 |
| mkks | 0.185 | gatm | -0.047 |
| CU499336.2 | 0.173 | apoa1b | -0.044 |
| zgc:174696 | 0.169 | mat1a | -0.044 |
| txlnbb | 0.161 | agxtb | -0.043 |
| si:dkey-19b23.14 | 0.156 | gapdh | -0.042 |
| FP101875.2 | 0.154 | afp4 | -0.041 |
| nexn | 0.154 | kng1 | -0.040 |
| zgc:194398 | 0.151 | si:dkey-16p21.8 | -0.040 |
| otofa | 0.150 | grhprb | -0.039 |
| rgl3a | 0.147 | pgm1 | -0.039 |
| entpd5b | 0.147 | serpinf2b | -0.039 |
| CABZ01073083.1 | 0.146 | serpinc1 | -0.038 |
| tfap2c | 0.145 | cpn1 | -0.038 |
| en1b | 0.137 | zgc:123103 | -0.037 |
| SRCIN1 | 0.137 | apoc2 | -0.037 |
| myod1 | 0.137 | f2 | -0.037 |
| adad2 | 0.136 | apoa2 | -0.037 |
| XLOC-026004 | 0.136 | tram1 | -0.037 |
| LOC100537874 | 0.134 | ppdpfa | -0.037 |
| LOC103910431 | 0.134 | cyp4v8 | -0.037 |
| C12orf75 | 0.133 | fabp10a | -0.037 |
| lmx1bb | 0.131 | serpina1 | -0.037 |
| LOC100330186 | 0.130 | c8g | -0.037 |
| XLOC-043918 | 0.127 | apoa4b.1 | -0.036 |
| LOC798089 | 0.126 | fbp1b | -0.036 |
| fbp2 | 0.124 | cx32.3 | -0.036 |
| zgc:112163 | 0.124 | fga | -0.036 |
| TESK1 | 0.123 | ttc36 | -0.036 |
| si:ch211-66i15.5 | 0.123 | si:ch73-361h17.1 | -0.036 |
| si:dkey-211g8.5 | 0.121 | nipsnap3a | -0.036 |