YJU2 splicing factor homolog
ZFIN 
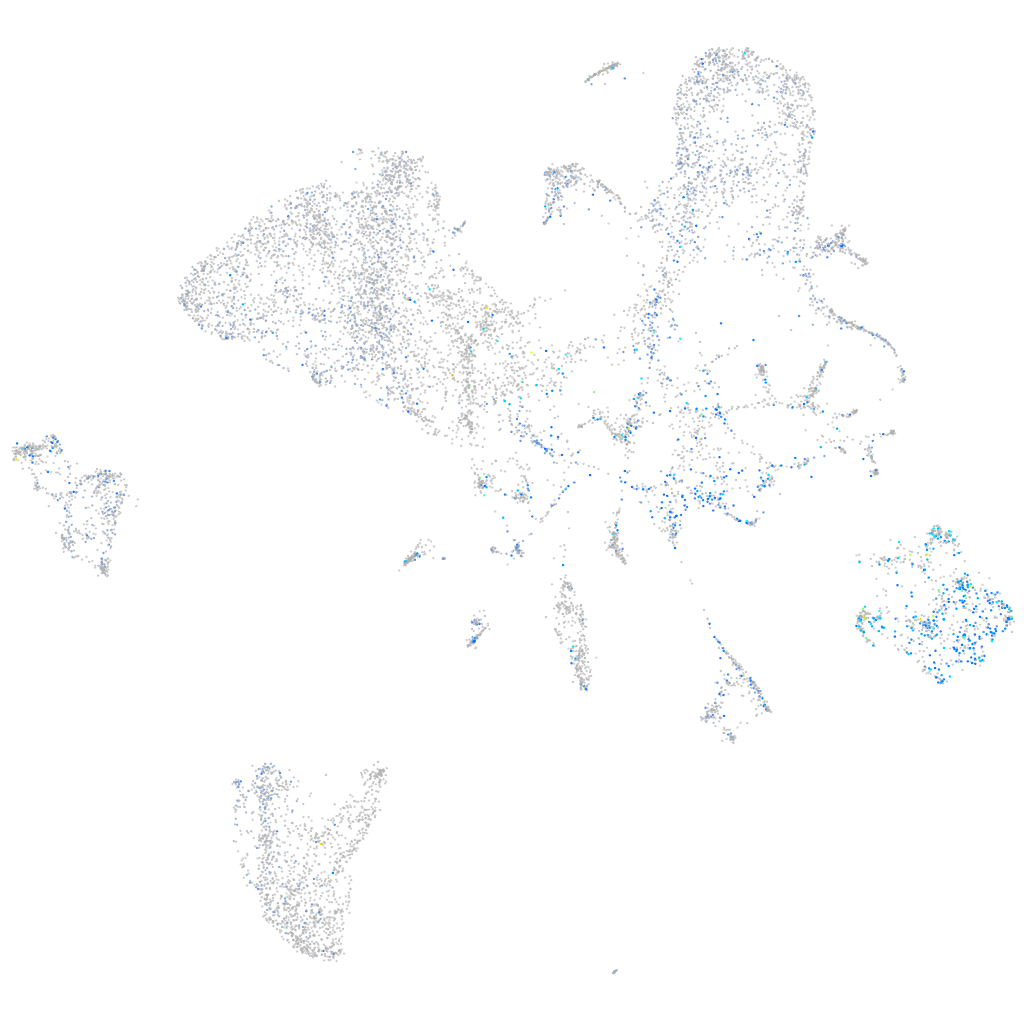
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| marcksb | 0.305 | rpl37 | -0.217 |
| syncrip | 0.297 | rps10 | -0.201 |
| hmga1a | 0.297 | gamt | -0.189 |
| hspb1 | 0.291 | zgc:114188 | -0.187 |
| si:ch211-152c2.3 | 0.290 | gapdh | -0.178 |
| seta | 0.288 | gatm | -0.175 |
| akap12b | 0.288 | ahcy | -0.170 |
| ilf3b | 0.286 | bhmt | -0.165 |
| si:ch73-281n10.2 | 0.286 | rps17 | -0.163 |
| hnrnpabb | 0.285 | zgc:92744 | -0.160 |
| hnrnpa1b | 0.284 | nupr1b | -0.157 |
| cx43.4 | 0.284 | zgc:158463 | -0.150 |
| nucks1a | 0.283 | apoa1b | -0.148 |
| cbx3a | 0.281 | mat1a | -0.147 |
| hdac1 | 0.280 | agxtb | -0.146 |
| anp32b | 0.280 | gpx4a | -0.146 |
| hnrnpa0b | 0.280 | eno3 | -0.145 |
| snrnp70 | 0.279 | pnp4b | -0.145 |
| anp32a | 0.279 | apoa2 | -0.141 |
| rbm4.3 | 0.278 | fbp1b | -0.140 |
| acin1a | 0.278 | aqp12 | -0.139 |
| hdgfl2 | 0.277 | dap | -0.139 |
| hnrnpaba | 0.277 | sod1 | -0.137 |
| setb | 0.277 | eef1da | -0.137 |
| hnrnpa1a | 0.276 | nme2b.1 | -0.136 |
| khdrbs1a | 0.276 | pklr | -0.136 |
| hmgb2a | 0.275 | apoa4b.1 | -0.135 |
| snrpd3l | 0.274 | afp4 | -0.134 |
| ncl | 0.274 | abat | -0.131 |
| ppig | 0.274 | rbp4 | -0.130 |
| smc1al | 0.273 | grhprb | -0.130 |
| hnrnpub | 0.273 | aldob | -0.128 |
| marcksl1b | 0.272 | apoc2 | -0.127 |
| top1l | 0.272 | suclg1 | -0.127 |
| si:ch211-51e12.7 | 0.270 | tpi1b | -0.127 |