YY1 associated factor 2
ZFIN 
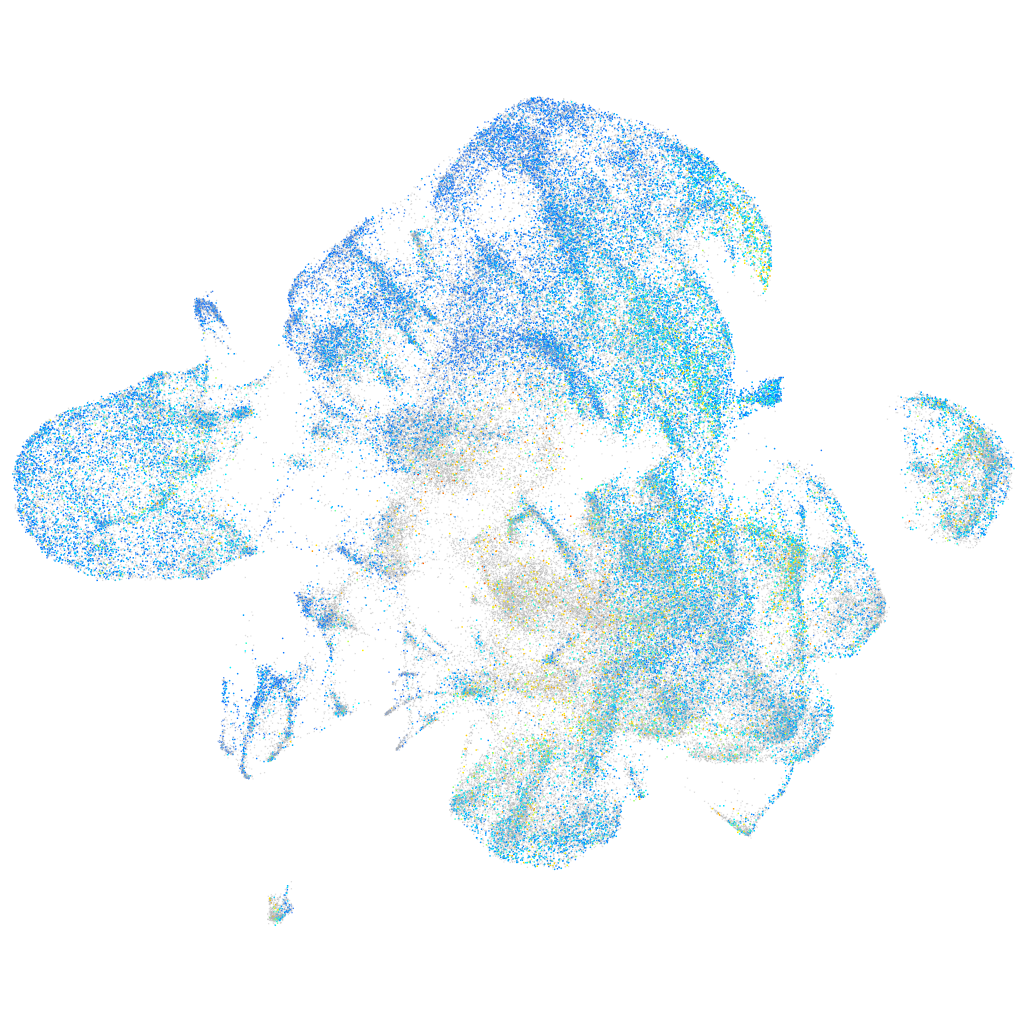
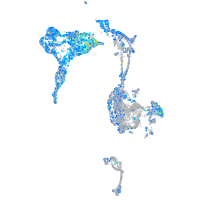
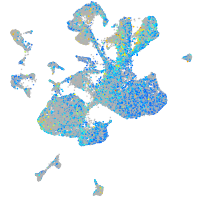
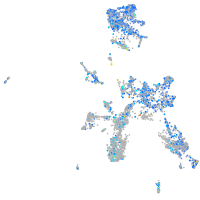
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ebf2 | 0.161 | ptprga | -0.059 |
| dlb | 0.152 | COX3 | -0.047 |
| sox11b | 0.148 | chd3 | -0.046 |
| hes6 | 0.146 | apoa2 | -0.046 |
| tp53inp2 | 0.142 | islr2 | -0.044 |
| CU634008.1 | 0.140 | xpr1a | -0.044 |
| hnrnpaba | 0.135 | apoa1b | -0.042 |
| LOC798783 | 0.135 | mt-atp6 | -0.042 |
| si:ch211-222l21.1 | 0.132 | cspg5a | -0.041 |
| cdkn1ca | 0.128 | si:ch73-127m5.1 | -0.040 |
| nhlh2 | 0.127 | syt1a | -0.039 |
| insm1a | 0.127 | aplp1 | -0.039 |
| notch1a | 0.126 | zgc:158463 | -0.038 |
| hmgn6 | 0.125 | gabrg2 | -0.038 |
| khdrbs1a | 0.122 | gabrb4 | -0.038 |
| cirbpb | 0.121 | pcdh7b | -0.038 |
| her13 | 0.120 | prss1 | -0.038 |
| rtca | 0.119 | myt1la | -0.037 |
| myt1b | 0.118 | pvalb2 | -0.037 |
| ptmab | 0.117 | sv2a | -0.037 |
| smad7 | 0.115 | slc6a1a | -0.036 |
| hdac1 | 0.114 | edil3a | -0.036 |
| gadd45gb.1 | 0.113 | prss59.2 | -0.036 |
| scrt2 | 0.112 | BX005003.1 | -0.035 |
| ubb | 0.111 | adcyap1r1a | -0.035 |
| smarce1 | 0.111 | pmp22a | -0.035 |
| dla | 0.110 | scg2b | -0.035 |
| syncrip | 0.110 | atp1a3a | -0.035 |
| gdf11 | 0.110 | nrxn2a | -0.035 |
| erh | 0.110 | ctrb1 | -0.035 |
| hnrnpabb | 0.108 | rtn1b | -0.034 |
| lima1a | 0.107 | nrxn1a | -0.034 |
| h3f3d | 0.107 | nlgn2b | -0.034 |
| sumo3a | 0.107 | maptb | -0.034 |
| hmga1a | 0.106 | pvalb1 | -0.034 |