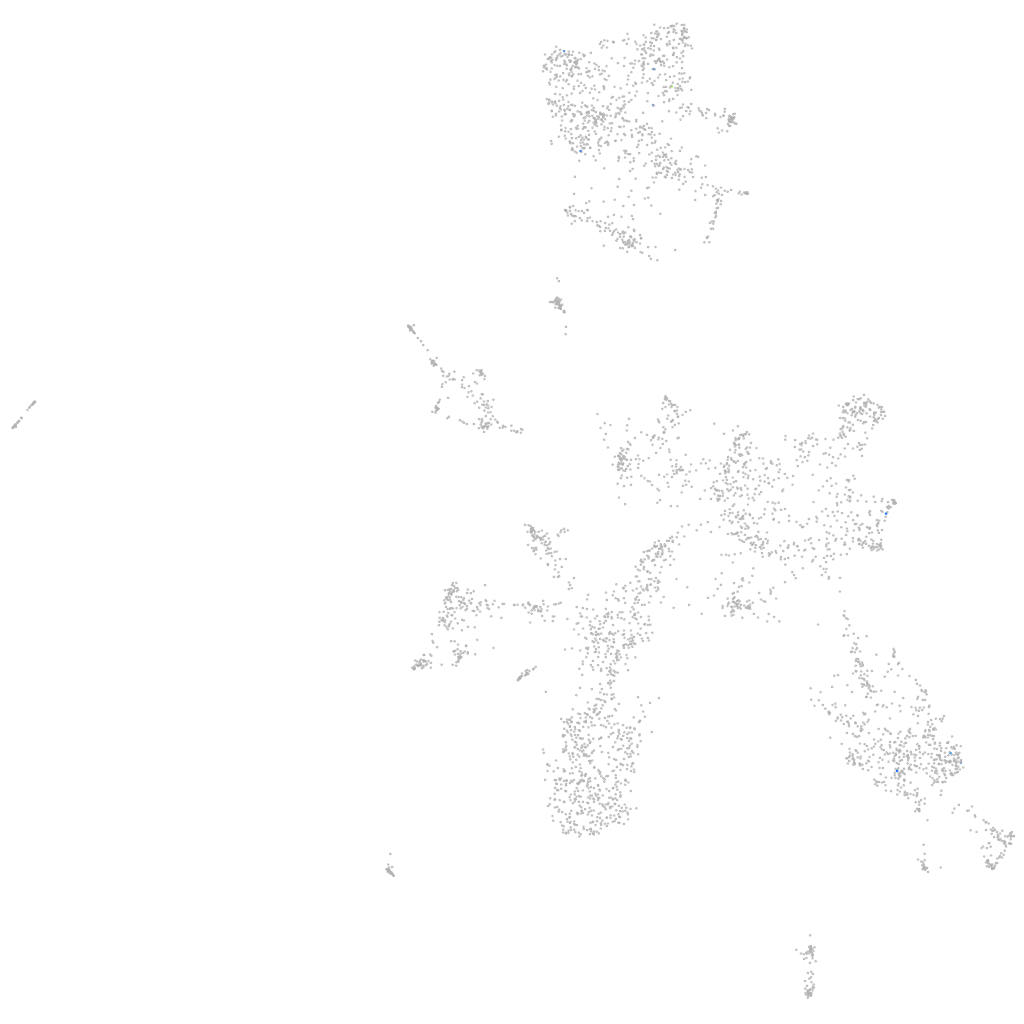
Other cell groups


all cells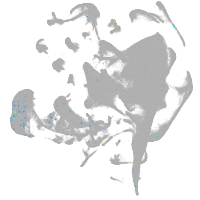 |
blastomeres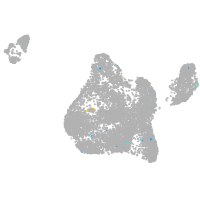 |
PGCs |
endoderm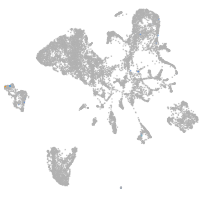 |
axial mesoderm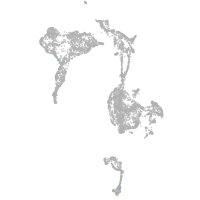 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 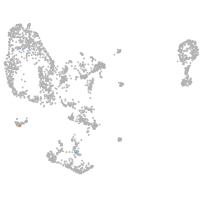 |
neural |
spinal cord / glia |
eye |
taste / olfactory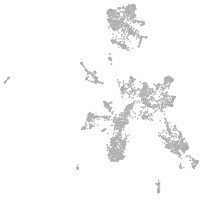 |
otic / lateral line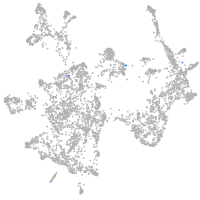 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:158432 | 0.632 | mt-co1 | -0.043 |
| slc26a3.2 | 0.625 | cox8a | -0.037 |
| nr5a5 | 0.613 | mt-co2 | -0.037 |
| myct1a | 0.534 | psmd3 | -0.034 |
| CABZ01001434.1 | 0.469 | CABZ01072614.1 | -0.029 |
| sult1st2 | 0.463 | pabpc1a | -0.029 |
| zgc:153968 | 0.385 | sub1a | -0.029 |
| XLOC-041297 | 0.382 | rab2a | -0.028 |
| ace2 | 0.348 | calm3b | -0.027 |
| fybb | 0.348 | NC-002333.4 | -0.026 |
| si:ch211-106h4.5 | 0.347 | gnb1b | -0.026 |
| irf4b | 0.320 | ywhaqb | -0.026 |
| zgc:77748 | 0.319 | calm2a | -0.025 |
| si:ch73-389b16.1 | 0.317 | elavl3 | -0.025 |
| LOC101886275 | 0.310 | hnrnpub | -0.025 |
| slc16a4 | 0.309 | eef1db | -0.025 |
| nkx1.2lb | 0.299 | calm1a | -0.024 |
| ins | 0.293 | mt-cyb | -0.024 |
| BX548061.1 | 0.291 | cox5ab | -0.024 |
| dgat2 | 0.278 | atrx | -0.023 |
| cers3a | 0.277 | hmgb3a | -0.023 |
| LOC100537368 | 0.269 | si:rp71-45k5.4 | -0.023 |
| myoz1a | 0.249 | atp5meb | -0.023 |
| cldn15a | 0.246 | nucks1b | -0.023 |
| serpinc1 | 0.241 | btf3l4 | -0.023 |
| hoxa9b | 0.236 | ammecr1 | -0.023 |
| parp9 | 0.230 | psma1 | -0.022 |
| rsad1 | 0.227 | sap30l | -0.022 |
| fga | 0.225 | tmsb | -0.022 |
| bin2b | 0.221 | ddx23 | -0.021 |
| six5 | 0.212 | mlf2 | -0.021 |
| si:dkey-262g12.12 | 0.209 | si:dkey-276j7.1 | -0.021 |
| LOC100329560 | 0.204 | srsf10b | -0.021 |
| XLOC-017188 | 0.204 | fip1l1b | -0.021 |
| RAB19 | 0.204 | psmc6 | -0.021 |