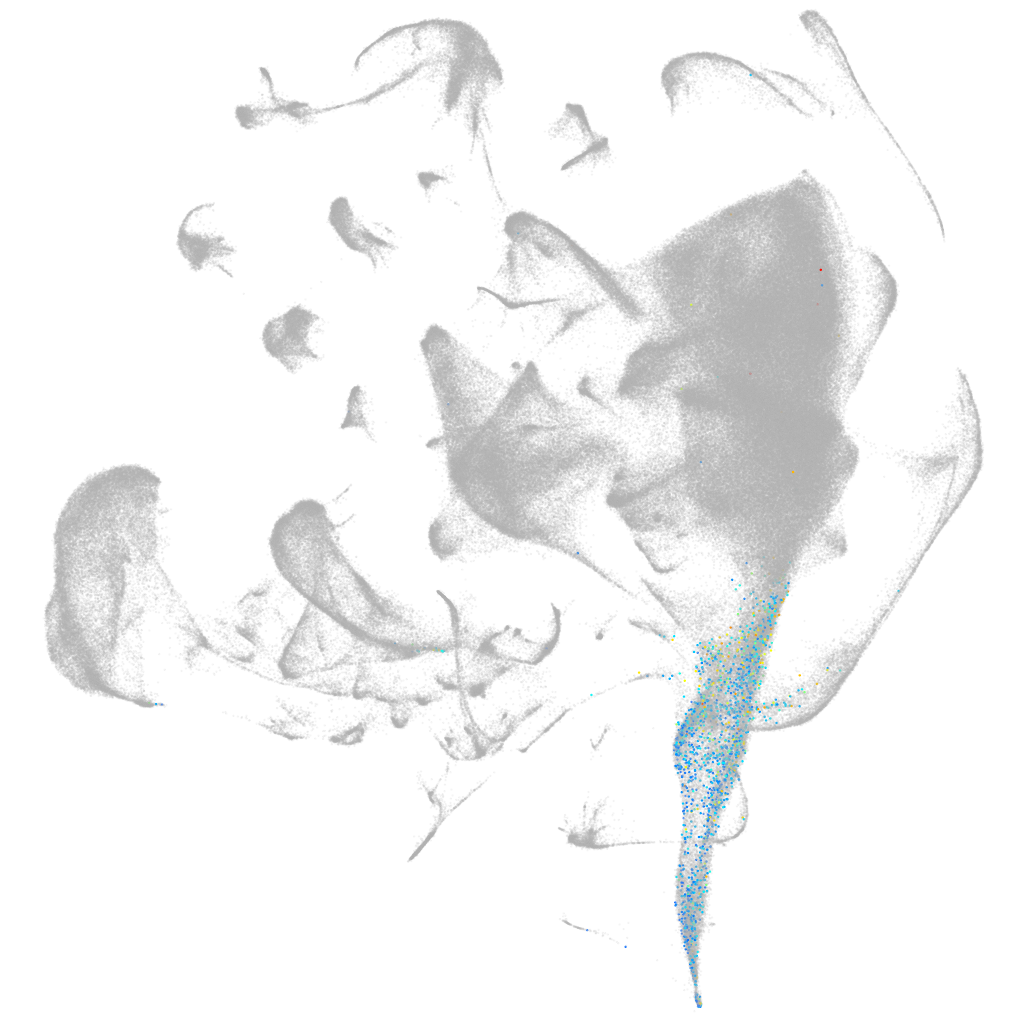
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-152c2.3 | 0.147 | si:ch1073-429i10.3.1 | -0.071 |
| hspb1 | 0.146 | calm1a | -0.065 |
| stm | 0.142 | ccni | -0.061 |
| si:dkey-66i24.9 | 0.141 | gapdhs | -0.058 |
| pou5f3 | 0.136 | tuba1c | -0.057 |
| s100a1 | 0.134 | marcksl1a | -0.057 |
| fbl | 0.132 | gpm6aa | -0.055 |
| zmp:0000000624 | 0.132 | hbbe1.3 | -0.055 |
| bms1 | 0.130 | rnasekb | -0.055 |
| COX7A2 | 0.130 | zgc:56493 | -0.054 |
| dkc1 | 0.130 | hbae3 | -0.053 |
| sp5l | 0.129 | rtn1a | -0.053 |
| nop58 | 0.127 | atp6v0cb | -0.052 |
| LOC108190024 | 0.124 | atp6v1e1b | -0.051 |
| npm1a | 0.124 | atp6v1g1 | -0.051 |
| polr3gla | 0.124 | krt4 | -0.051 |
| add3b | 0.123 | stmn1b | -0.051 |
| ddx18 | 0.123 | hbae1.1 | -0.050 |
| gar1 | 0.122 | pvalb1 | -0.050 |
| hnrnpa1b | 0.122 | cyt1 | -0.049 |
| ppig | 0.122 | wu:fb55g09 | -0.049 |
| gnl3 | 0.121 | cct2 | -0.049 |
| kri1 | 0.121 | gng3 | -0.048 |
| ncl | 0.120 | pvalb2 | -0.048 |
| vrtn | 0.120 | ckbb | -0.047 |
| lig1 | 0.118 | gnb1a | -0.046 |
| vox | 0.118 | gpm6ab | -0.046 |
| ing5b | 0.117 | hbbe1.1 | -0.046 |
| nop2 | 0.117 | sncb | -0.046 |
| nop56 | 0.117 | cyt1l | -0.045 |
| polr2gl | 0.117 | elavl3 | -0.045 |
| dnmt3bb.2 | 0.116 | vamp2 | -0.045 |
| rbmx2 | 0.116 | fabp3 | -0.044 |
| rrp15 | 0.116 | gabarapl2 | -0.044 |
| si:dkey-92i17.2 | 0.116 | prdx2 | -0.044 |