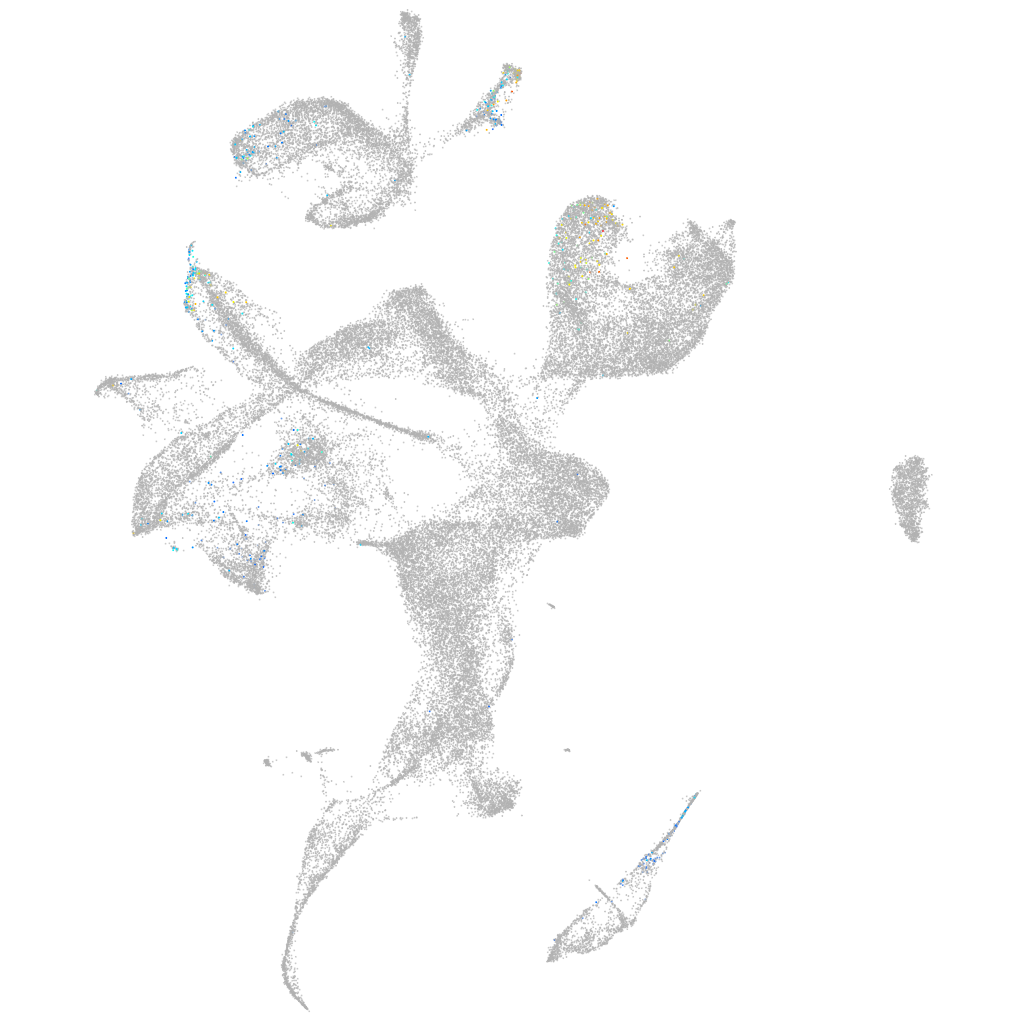
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| eno1a | 0.141 | rps20 | -0.104 |
| kcnh2a | 0.136 | rplp2l | -0.100 |
| tnr | 0.132 | rpl11 | -0.098 |
| ntng1a | 0.131 | rps12 | -0.096 |
| CR361564.1 | 0.124 | rps23 | -0.096 |
| slc17a6b | 0.123 | rpl26 | -0.094 |
| btbd17b | 0.121 | rpl36a | -0.093 |
| si:ch211-151p13.8 | 0.121 | rpl39 | -0.093 |
| adcy3a | 0.120 | rpl13a | -0.093 |
| si:ch73-193i22.1 | 0.116 | rps15a | -0.092 |
| gria3a | 0.114 | rps7 | -0.092 |
| kcnj2b | 0.110 | rps14 | -0.092 |
| aldocb | 0.109 | rpsa | -0.092 |
| slc2a3a | 0.108 | rps27.1 | -0.091 |
| BX897741.1 | 0.108 | rps13 | -0.091 |
| grin1a | 0.107 | rplp1 | -0.091 |
| btbd17a | 0.107 | rpl12 | -0.090 |
| slc4a5a | 0.104 | rpl10 | -0.089 |
| si:ch211-153l6.6 | 0.104 | rps21 | -0.089 |
| atp2b3b | 0.104 | rpl8 | -0.089 |
| gapdhs | 0.103 | rpl36 | -0.088 |
| edil3a | 0.103 | rplp0 | -0.088 |
| lrrc4.1 | 0.103 | rpl13 | -0.088 |
| opn9 | 0.102 | rpl23 | -0.087 |
| kcnj14 | 0.099 | rpl18a | -0.087 |
| rprmb | 0.099 | rps5 | -0.086 |
| si:dkey-16p21.8 | 0.099 | rpl27 | -0.086 |
| vsnl1b | 0.096 | hmga1a | -0.086 |
| grik1a | 0.096 | rps9 | -0.086 |
| ube2ql1 | 0.095 | rps27a | -0.086 |
| CU861453.1 | 0.094 | rpl10a | -0.086 |
| amigo1 | 0.093 | rpl7 | -0.086 |
| slc4a8 | 0.093 | si:dkey-151g10.6 | -0.086 |
| LOC108179567 | 0.091 | rpl19 | -0.085 |
| cx52.9 | 0.091 | rps4x | -0.085 |