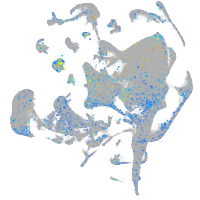
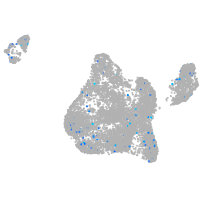
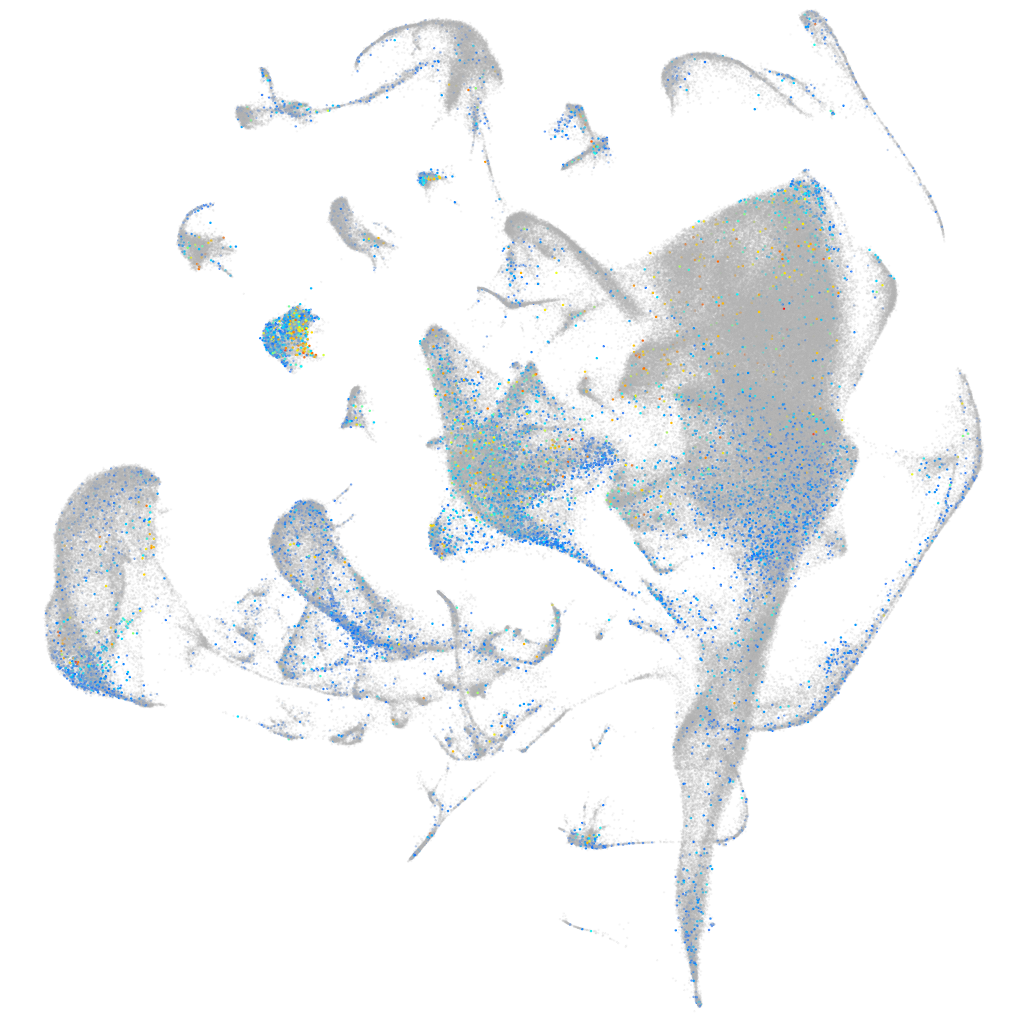Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pecam1 | 0.112 | tuba1c | -0.055 |
| plvapb | 0.111 | gpm6aa | -0.050 |
| tpm4a | 0.111 | elavl3 | -0.049 |
| clec14a | 0.110 | si:ch211-137a8.4 | -0.049 |
| cdh5 | 0.109 | tuba1a | -0.048 |
| si:dkeyp-97a10.2 | 0.108 | ckbb | -0.045 |
| kdr | 0.106 | rtn1a | -0.044 |
| kdrl | 0.106 | tmsb | -0.044 |
| tie1 | 0.106 | hmgb1b | -0.043 |
| myct1a | 0.105 | stmn1b | -0.043 |
| si:ch211-145b13.6 | 0.102 | nova2 | -0.042 |
| krt8 | 0.098 | si:dkey-276j7.1 | -0.041 |
| rasip1 | 0.097 | epb41a | -0.040 |
| she | 0.097 | hes6 | -0.039 |
| etv2 | 0.096 | myt1a | -0.039 |
| si:dkey-261h17.1 | 0.096 | marcksb | -0.038 |
| pmp22a | 0.095 | chd4a | -0.037 |
| ecscr | 0.095 | myt1b | -0.037 |
| krt18a.1 | 0.094 | mdkb | -0.036 |
| ramp2 | 0.092 | sox11b | -0.036 |
| si:ch73-334d15.2 | 0.092 | scrt2 | -0.035 |
| scarf1 | 0.090 | dlb | -0.034 |
| mcamb | 0.088 | rnasekb | -0.034 |
| adgrl4 | 0.086 | CU634008.1 | -0.034 |
| yrk | 0.086 | cadm3 | -0.033 |
| egfl7 | 0.085 | CU467822.1 | -0.033 |
| exoc3l2a | 0.085 | insm1a | -0.033 |
| fstl1b | 0.085 | tubb5 | -0.033 |
| emp2 | 0.084 | zc4h2 | -0.033 |
| fli1a | 0.084 | hmgb3a | -0.032 |
| flt1 | 0.084 | tmed4 | -0.032 |
| gpr182 | 0.084 | tmpob | -0.032 |
| col4a1 | 0.083 | fabp7a | -0.031 |
| fgd5a | 0.083 | si:dkey-56m19.5 | -0.031 |
| myct1b | 0.083 | LOC100537384 | -0.031 |