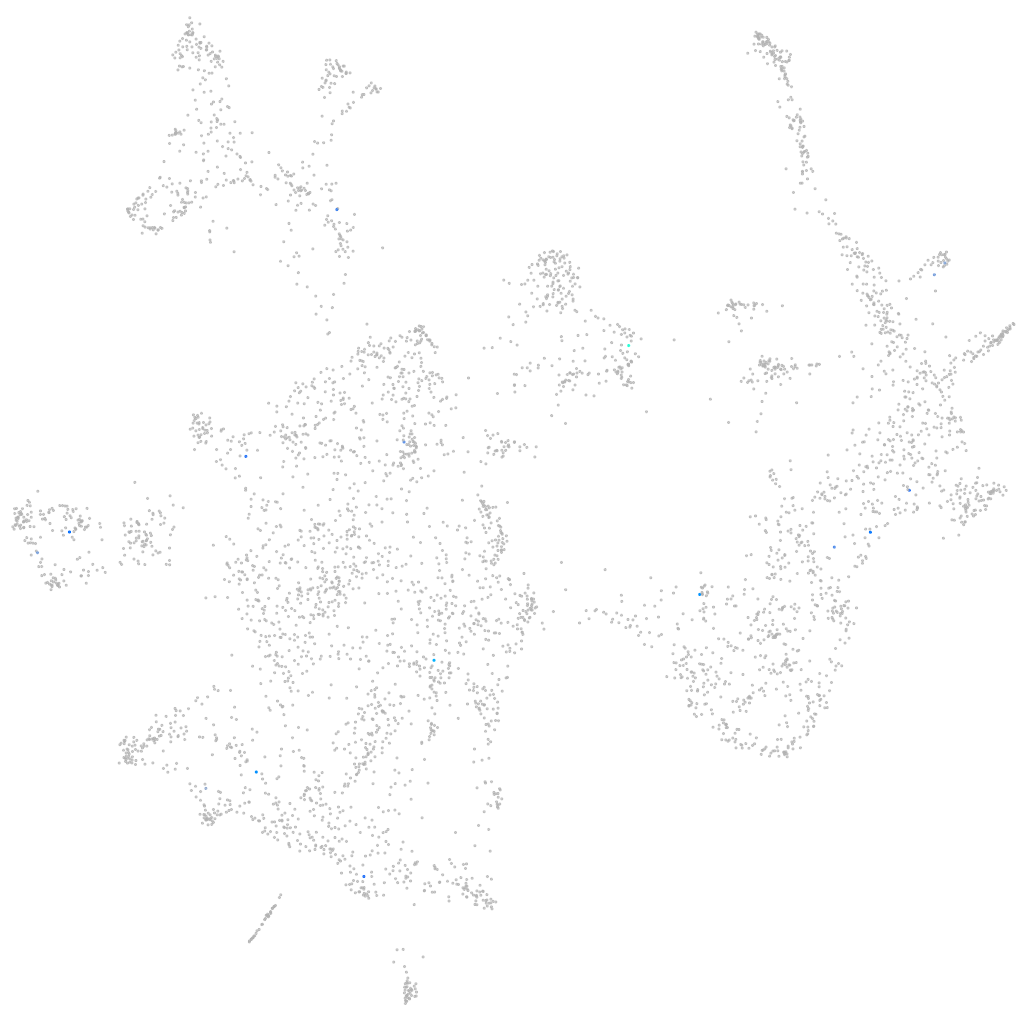
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural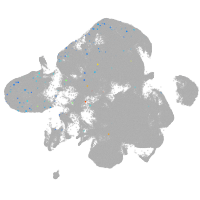 |
spinal cord / glia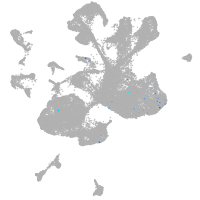 |
eye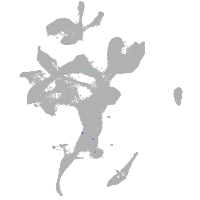 |
taste / olfactory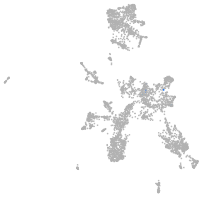 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-74m13.1 | 0.562 | h1f0 | -0.044 |
| CABZ01057381.1 | 0.474 | eno1a | -0.029 |
| asip1 | 0.420 | anp32a | -0.028 |
| hbl4 | 0.407 | itm2ba | -0.028 |
| znf1061 | 0.381 | six1b | -0.028 |
| si:ch211-227m13.1 | 0.319 | zgc:92606 | -0.027 |
| XLOC-023348 | 0.318 | mdh2 | -0.027 |
| dgat2 | 0.308 | atp5f1b | -0.026 |
| esrrgb | 0.297 | ccnl1a | -0.025 |
| rxfp3.2b | 0.295 | NDUFB1 | -0.025 |
| BX004779.2 | 0.285 | rab28 | -0.025 |
| tat | 0.276 | alas1 | -0.024 |
| LOC108181502 | 0.274 | cox6b1 | -0.024 |
| si:ch211-133l5.7 | 0.269 | tm2d3 | -0.024 |
| itga11a | 0.253 | dicer1 | -0.024 |
| CABZ01092907.1 | 0.236 | sox10 | -0.024 |
| znf1037 | 0.233 | ndufs8a | -0.024 |
| LO017821.1 | 0.232 | COX3 | -0.024 |
| LOC110439626 | 0.232 | cst14b.1 | -0.024 |
| BX927130.2 | 0.232 | cox5aa | -0.024 |
| ins | 0.229 | ndufa3 | -0.023 |
| CU693446.1 | 0.225 | nipsnap2 | -0.023 |
| tpbga | 0.203 | atp6v1ba | -0.023 |
| f13a1b | 0.201 | tktb | -0.023 |
| hpdb | 0.201 | tufm | -0.023 |
| guk1b | 0.198 | ckbb | -0.023 |
| pax1b | 0.197 | tbl1xr1a | -0.022 |
| adgra2 | 0.196 | ndufs7 | -0.022 |
| BX248501.1 | 0.192 | ckmt1 | -0.022 |
| arhgap31 | 0.191 | phrf1 | -0.022 |
| trim63a | 0.190 | pnrc2 | -0.022 |
| twist2 | 0.189 | psmd9 | -0.022 |
| mir199-3 | 0.185 | sod2 | -0.022 |
| itln1l | 0.185 | ralbp1 | -0.022 |
| LOC108183534 | 0.184 | stox1 | -0.022 |