Other cell groups


all cells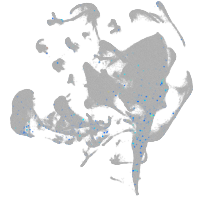 |
blastomeres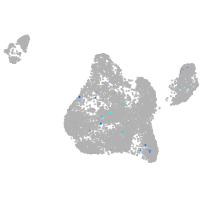 |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 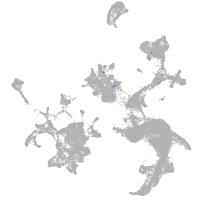 |
pronephros  |
neural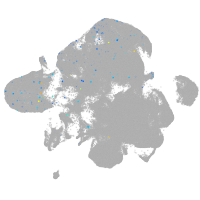 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC100148254 | 0.126 | ckbb | -0.040 |
| LOC108181391 | 0.118 | rtn1a | -0.035 |
| LOC101886091 | 0.091 | gpm6ab | -0.032 |
| CU499311.1 | 0.088 | elavl3 | -0.031 |
| BX248418.1 | 0.086 | si:dkeyp-75h12.5 | -0.030 |
| BX537274.1 | 0.079 | calm1a | -0.029 |
| LOC110439463 | 0.072 | pvalb1 | -0.029 |
| nkd3l | 0.071 | myt1b | -0.028 |
| dut | 0.070 | gdi1 | -0.028 |
| BX005442.2 | 0.067 | gpm6aa | -0.027 |
| gig2i | 0.066 | slc1a2b | -0.027 |
| LOC108190395 | 0.065 | rnasekb | -0.027 |
| mki67 | 0.064 | tmsb | -0.027 |
| ccna2 | 0.063 | stmn1b | -0.027 |
| pcna | 0.063 | CR383676.1 | -0.026 |
| cks1b | 0.063 | pvalb2 | -0.026 |
| cdca5 | 0.062 | ywhah | -0.026 |
| dek | 0.062 | COX3 | -0.025 |
| LOC101885088 | 0.060 | hbbe1.3 | -0.025 |
| stmn1a | 0.060 | stx1b | -0.025 |
| AL954322.3 | 0.059 | ube2d4 | -0.025 |
| aurkb | 0.059 | calr | -0.025 |
| tmc2a | 0.059 | scrt2 | -0.024 |
| RF00070 | 0.059 | fez1 | -0.024 |
| esco2 | 0.058 | aldocb | -0.024 |
| rrm1 | 0.057 | vamp2 | -0.024 |
| RF00099 | 0.057 | cadm4 | -0.024 |
| fbxo5 | 0.057 | hbae3 | -0.024 |
| BX569796.1 | 0.057 | atp6v1g1 | -0.024 |
| hmgb2a | 0.057 | gng3 | -0.024 |
| si:dkey-9p20.1 | 0.056 | gap43 | -0.024 |
| si:ch211-232d10.3 | 0.056 | elavl4 | -0.024 |
| smc2 | 0.056 | dpysl3 | -0.024 |
| LOC100330864 | 0.056 | gapdhs | -0.023 |
| zgc:110540 | 0.056 | si:ch211-133n4.4 | -0.023 |




