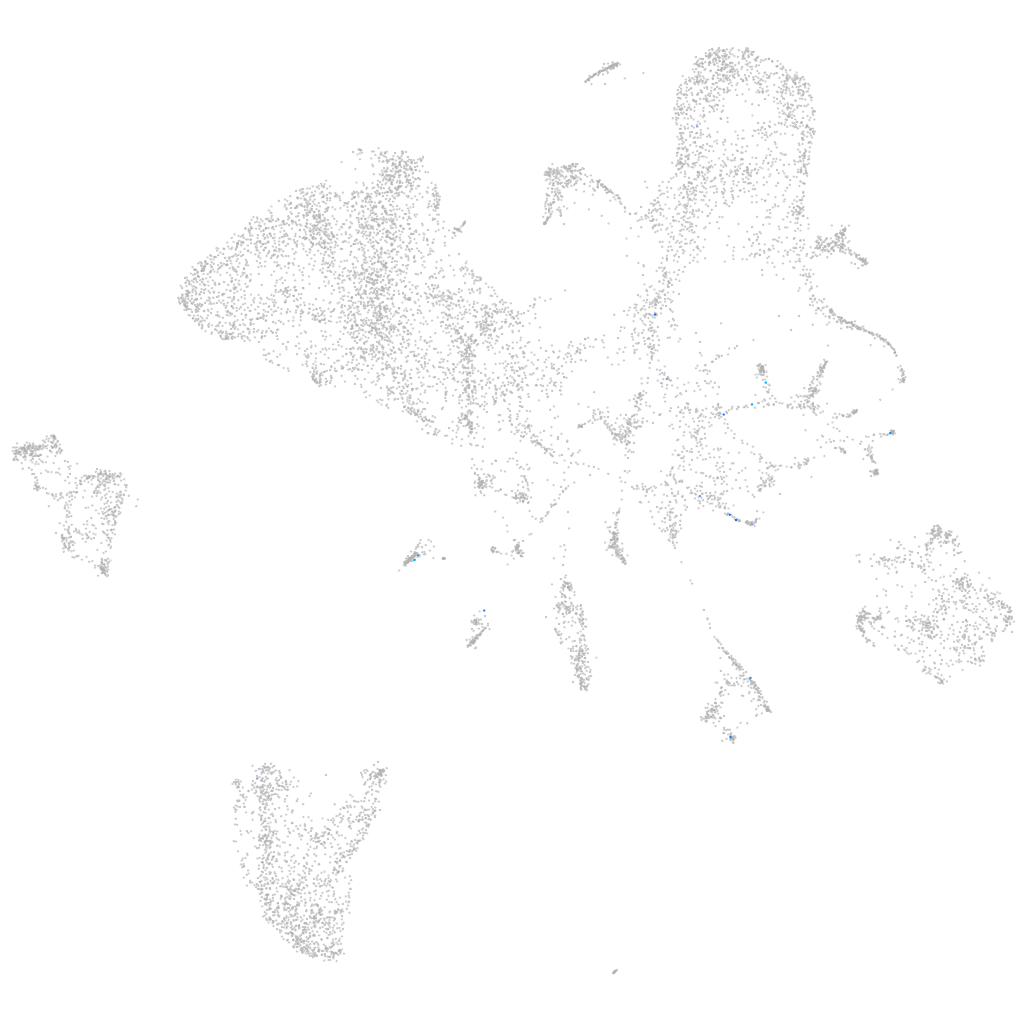
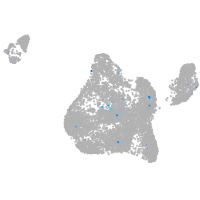
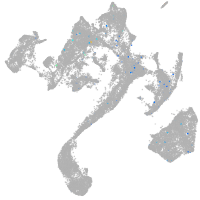
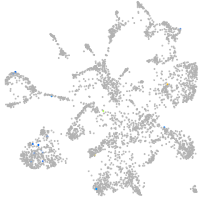
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC100537874 | 0.363 | bhmt | -0.037 |
| si:ch211-66i15.5 | 0.334 | apoc1 | -0.034 |
| si:dkey-211g8.5 | 0.329 | gamt | -0.034 |
| CR936974.1 | 0.317 | gapdh | -0.033 |
| irbpl | 0.315 | apoa1b | -0.033 |
| LOC101882178 | 0.311 | apoa2 | -0.032 |
| BX927369.1 | 0.279 | gatm | -0.032 |
| prokr1b | 0.279 | grhprb | -0.031 |
| aifm5 | 0.265 | agxtb | -0.031 |
| CR589943.1 | 0.244 | ahcy | -0.030 |
| crygs3 | 0.220 | zgc:123103 | -0.029 |
| LOC103910526 | 0.220 | rbp2b | -0.029 |
| LOC101886558 | 0.194 | serpina1l | -0.029 |
| si:ch211-284e13.14 | 0.182 | serpina1 | -0.029 |
| cdkn2a/b | 0.181 | fabp10a | -0.029 |
| celsr3 | 0.179 | cfhl4 | -0.029 |
| chpfb | 0.178 | apom | -0.028 |
| adcy1a | 0.175 | mat1a | -0.028 |
| XLOC-034550 | 0.171 | fetub | -0.028 |
| fyba | 0.160 | fgb | -0.028 |
| CR812469.1 | 0.157 | ces2 | -0.028 |
| rsad2 | 0.155 | ambp | -0.028 |
| EGFLAM | 0.151 | LOC110437731 | -0.028 |
| CABZ01070579.1 | 0.149 | hpda | -0.027 |
| xkr5a | 0.146 | shmt1 | -0.027 |
| LOC101887079 | 0.144 | agxta | -0.027 |
| AP3B2 | 0.143 | uox | -0.027 |
| si:dkey-210j14.3 | 0.143 | f2 | -0.027 |
| CU469524.2 | 0.130 | rbp4 | -0.027 |
| si:ch211-156l18.7 | 0.124 | fga | -0.027 |
| myoz2b | 0.122 | ftcd | -0.027 |
| LOC103910514 | 0.120 | pnp4b | -0.027 |
| si:dkey-61p9.11 | 0.118 | hao1 | -0.027 |
| si:ch73-177h5.3 | 0.118 | ttr | -0.026 |
| npy7r | 0.118 | serpinc1 | -0.026 |