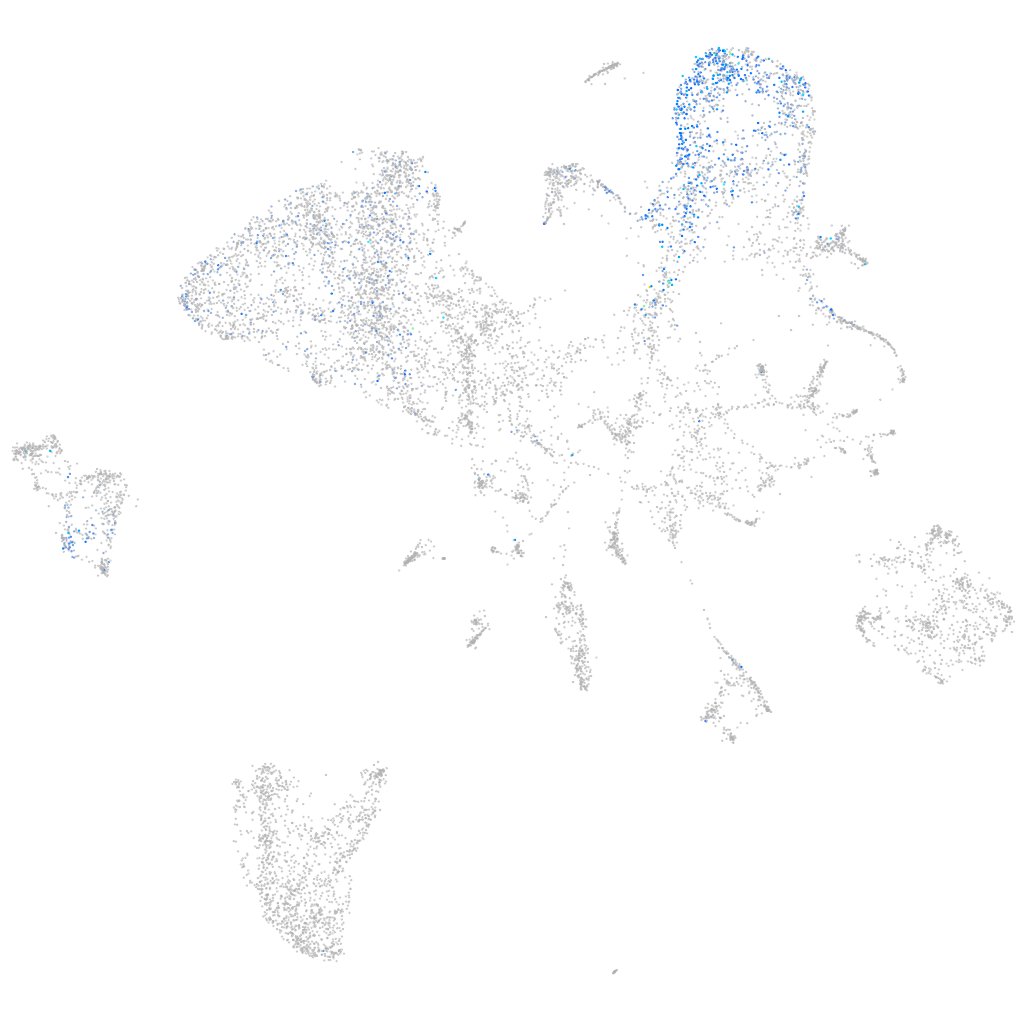
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aqp8a.2 | 0.459 | fabp3 | -0.142 |
| ace2 | 0.436 | krt18a.1 | -0.134 |
| cpo | 0.432 | aqp12 | -0.134 |
| slc15a1b | 0.429 | rtn1a | -0.132 |
| si:ch211-113d11.6 | 0.428 | bhmt | -0.125 |
| cd36 | 0.425 | marcksl1b | -0.117 |
| stoml3b | 0.418 | hmgb3a | -0.114 |
| chia.2 | 0.418 | cd81a | -0.112 |
| ldha | 0.416 | marcksb | -0.109 |
| wu:fa56d06 | 0.415 | qkia | -0.109 |
| lta4h | 0.412 | ctrl | -0.107 |
| mogat2 | 0.410 | ppdpfb | -0.106 |
| mansc1 | 0.405 | jpt1b | -0.105 |
| ada | 0.404 | zgc:112160 | -0.105 |
| rbp2a | 0.402 | fep15 | -0.103 |
| rtn4b | 0.402 | cel.1 | -0.103 |
| dhrs1 | 0.400 | apoda.2 | -0.103 |
| acsl5 | 0.399 | prss59.2 | -0.103 |
| slc6a19a.2 | 0.399 | CELA1 (1 of many) | -0.102 |
| slc6a19a.1 | 0.399 | cpa5 | -0.102 |
| zgc:153968 | 0.398 | prss59.1 | -0.102 |
| mbl2 | 0.398 | sycn.2 | -0.102 |
| zgc:103681 | 0.397 | pdia2 | -0.102 |
| ugt1a7 | 0.397 | prss1 | -0.101 |
| zgc:158846 | 0.395 | erp27 | -0.101 |
| meltf | 0.394 | ela2l | -0.101 |
| fabp1b.1 | 0.393 | ela3l | -0.101 |
| aoc1 | 0.393 | si:ch211-240l19.5 | -0.100 |
| chia.1 | 0.392 | cpb1 | -0.100 |
| atp1a1a.4 | 0.392 | ela2 | -0.100 |
| si:ch211-133l5.7 | 0.392 | ctrb1 | -0.100 |
| ndufa4 | 0.390 | ptmab | -0.100 |
| si:ch211-161h7.8 | 0.390 | cel.2 | -0.099 |
| dhrs13l1 | 0.387 | zgc:136461 | -0.099 |
| slc15a1a | 0.386 | fxyd1 | -0.098 |