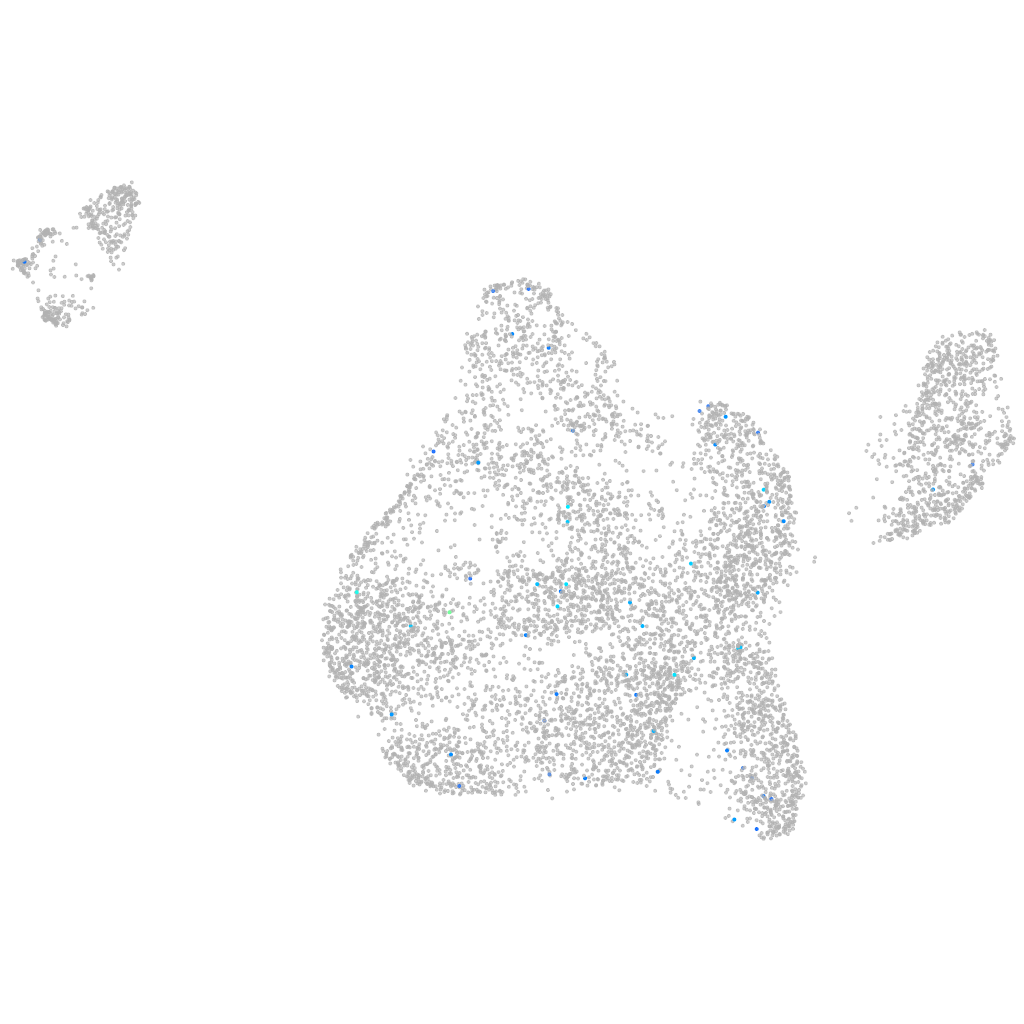
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm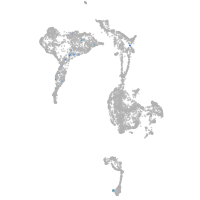 |
muscle 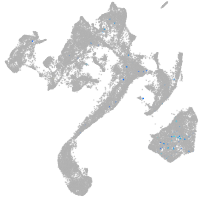 |
mural cells / non-skeletal muscle  |
mesenchyme 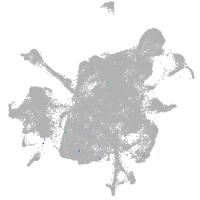 |
hematopoietic / vasculature 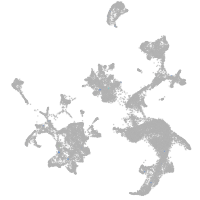 |
pronephros 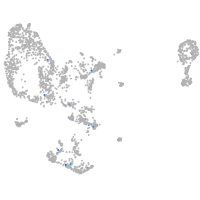 |
neural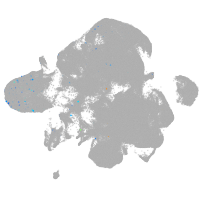 |
spinal cord / glia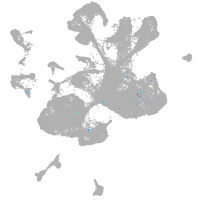 |
eye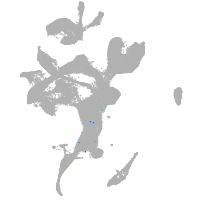 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gja3 | 0.169 | npc2 | -0.035 |
| prf1.5 | 0.151 | tmem30b | -0.034 |
| CR388421.1 | 0.149 | tram1 | -0.032 |
| srd5a2b | 0.145 | nap1l4a | -0.032 |
| LOC101885655 | 0.133 | fam117ab | -0.032 |
| slc6a8 | 0.131 | sf3b4 | -0.031 |
| agap2 | 0.131 | gra | -0.030 |
| si:ch73-190m4.1 | 0.130 | srrm1 | -0.030 |
| LOC103909069 | 0.129 | ctnna1 | -0.030 |
| FAT4 | 0.125 | mycn | -0.030 |
| si:dkey-152b24.8 | 0.120 | cuedc2 | -0.029 |
| si:ch211-120k19.1 | 0.120 | cat | -0.029 |
| ora4 | 0.116 | STMP1 | -0.029 |
| slc22a13a | 0.111 | zgc:77118 | -0.028 |
| LOC101886950 | 0.109 | snx3 | -0.028 |
| GUCY1A2 | 0.109 | ckbb | -0.028 |
| jac8 | 0.108 | mkrn4 | -0.028 |
| taar16a | 0.102 | cldnd | -0.028 |
| CR762475.1 | 0.100 | eef1da | -0.028 |
| ftr29 | 0.098 | zgc:161969 | -0.028 |
| LOC110438887 | 0.098 | pak2a | -0.028 |
| RF00056 | 0.098 | siva1 | -0.028 |
| XLOC-043072 | 0.097 | fam222bb | -0.028 |
| si:dkey-21h14.9 | 0.095 | lgmn | -0.028 |
| LOC110439557 | 0.092 | snrpc | -0.028 |
| LOC110437938 | 0.091 | top1l | -0.028 |
| socs7 | 0.091 | zgc:162025 | -0.027 |
| zgc:158263 | 0.091 | eif2s1a | -0.027 |
| prl | 0.091 | zgc:77650 | -0.027 |
| nr5a2 | 0.090 | uba5 | -0.027 |
| or137-3 | 0.089 | syncrip | -0.027 |
| sh3pxd2aa | 0.089 | ndufv1 | -0.027 |
| BX511034.6 | 0.087 | arl8 | -0.027 |
| gpr158a | 0.086 | si:dkey-42i9.8 | -0.027 |
| LOC101882181 | 0.086 | polr2gl | -0.027 |