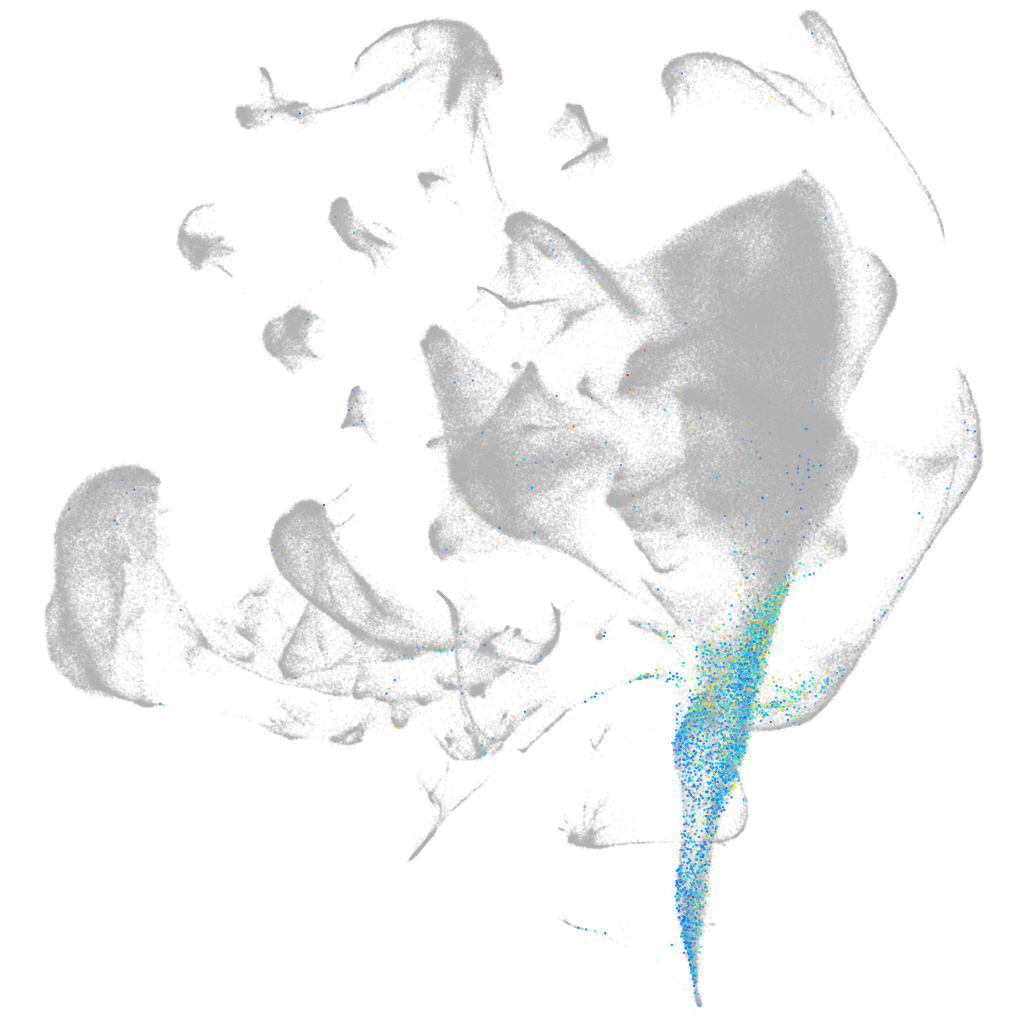
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.220 | si:ch1073-429i10.3.1 | -0.105 |
| si:ch211-152c2.3 | 0.220 | calm1a | -0.092 |
| zmp:0000000624 | 0.213 | ccni | -0.090 |
| dkc1 | 0.197 | marcksl1a | -0.086 |
| fbl | 0.196 | zgc:56493 | -0.086 |
| COX7A2 | 0.192 | gapdhs | -0.082 |
| nop58 | 0.192 | tuba1c | -0.081 |
| bms1 | 0.191 | hbbe1.3 | -0.079 |
| stm | 0.190 | gpm6aa | -0.078 |
| nop2 | 0.189 | cct2 | -0.077 |
| nr6a1a | 0.189 | rnasekb | -0.076 |
| gnl3 | 0.188 | hbae3 | -0.075 |
| npm1a | 0.187 | rtn1a | -0.075 |
| si:dkey-66i24.9 | 0.187 | atp6v1g1 | -0.074 |
| gar1 | 0.184 | atp6v1e1b | -0.073 |
| ddx18 | 0.183 | krt4 | -0.073 |
| ncl | 0.183 | pvalb1 | -0.073 |
| nop56 | 0.180 | ckbb | -0.072 |
| pprc1 | 0.178 | stmn1b | -0.072 |
| rbmx2 | 0.177 | wu:fb55g09 | -0.072 |
| ppig | 0.176 | atp6v0cb | -0.071 |
| lig1 | 0.175 | cyt1 | -0.071 |
| ebna1bp2 | 0.174 | fabp3 | -0.071 |
| kri1 | 0.174 | hbae1.1 | -0.071 |
| pes | 0.174 | gnb1a | -0.070 |
| rrp15 | 0.173 | pvalb2 | -0.070 |
| rsl1d1 | 0.173 | gng3 | -0.067 |
| hnrnpa1b | 0.171 | gpm6ab | -0.067 |
| mybbp1a | 0.170 | prdx2 | -0.066 |
| polr3gla | 0.168 | sncb | -0.065 |
| mphosph10 | 0.162 | cyt1l | -0.064 |
| ISCU | 0.160 | elavl3 | -0.064 |
| si:dkey-239h2.3 | 0.160 | gabarapb | -0.064 |
| rrp1 | 0.159 | gabarapl2 | -0.064 |
| shisa2a | 0.159 | tob1b | -0.064 |