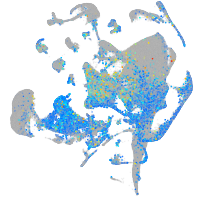
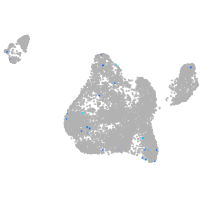
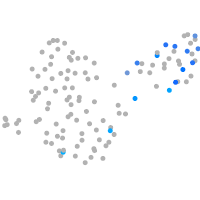
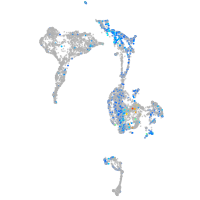
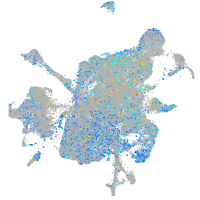
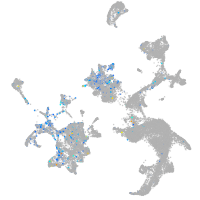
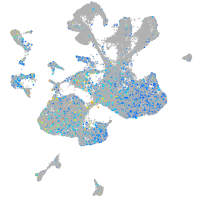
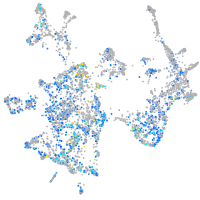
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01075068.1 | 0.076 | ckbb | -0.048 |
| eef1a1l1 | 0.073 | mia | -0.046 |
| lin28a | 0.072 | gpm6aa | -0.043 |
| zbtb16a | 0.068 | stmn1b | -0.042 |
| tmem88b | 0.064 | rtn1a | -0.042 |
| si:dkey-195m11.8 | 0.061 | acana | -0.042 |
| tpm4a | 0.058 | snorc | -0.041 |
| eef2b | 0.058 | elavl3 | -0.041 |
| si:dkey-42i9.4 | 0.055 | cnmd | -0.040 |
| serbp1a | 0.055 | tmsb | -0.040 |
| eif3ba | 0.055 | matn1 | -0.038 |
| si:ch211-286o17.1 | 0.054 | col2a1a | -0.037 |
| actb2 | 0.054 | stx1b | -0.037 |
| zfp36l1a | 0.053 | atp6v0cb | -0.036 |
| pmp22a | 0.053 | ndrg4 | -0.036 |
| cx43.4 | 0.053 | zgc:158423 | -0.035 |
| sdc4 | 0.052 | wisp3 | -0.035 |
| hspa8 | 0.051 | fgfbp2b | -0.035 |
| ptbp1a | 0.051 | itih1 | -0.035 |
| XLOC-042222 | 0.051 | elavl4 | -0.035 |
| qkia | 0.051 | ptgdsa | -0.035 |
| trim71 | 0.050 | mdkb | -0.034 |
| id3 | 0.050 | si:dkey-19b23.8 | -0.034 |
| selenoh | 0.049 | ppp1r1c | -0.034 |
| eif4ebp3l | 0.049 | otos | -0.034 |
| her6 | 0.048 | col9a1a | -0.033 |
| hnrnpa0l | 0.048 | fxyd1 | -0.033 |
| marcksl1a | 0.048 | cspg5a | -0.033 |
| ftr82 | 0.048 | gapdhs | -0.033 |
| mcm7 | 0.048 | si:dkey-6n6.1 | -0.033 |
| rps3a | 0.048 | sncb | -0.033 |
| arl4cb | 0.048 | epyc | -0.033 |
| igf2bp1 | 0.047 | tnxba | -0.032 |
| snrpb | 0.047 | emilin3a | -0.032 |
| CABZ01072614.1 | 0.047 | ucmab | -0.032 |