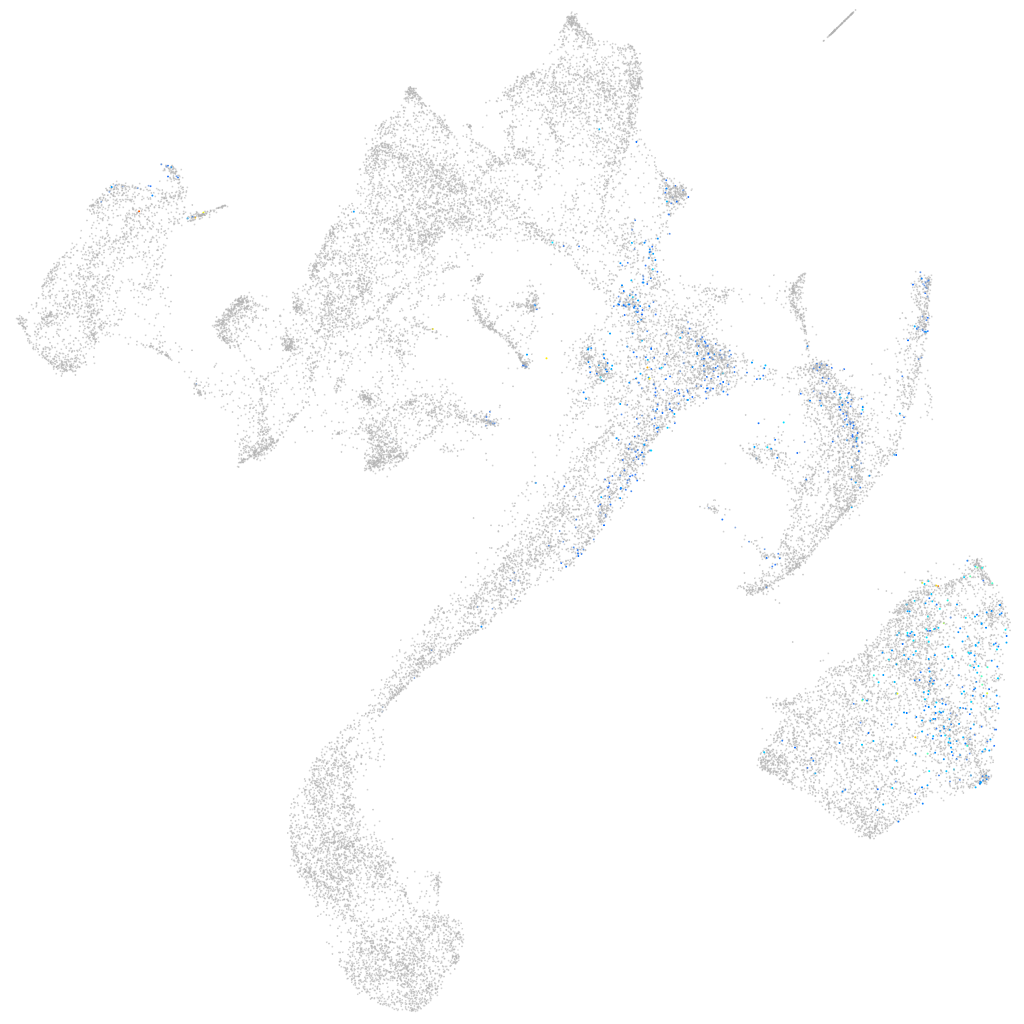
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX927258.1 | 0.160 | fabp3 | -0.112 |
| im:7138239 | 0.133 | rpl37 | -0.100 |
| ebna1bp2 | 0.131 | vdac3 | -0.100 |
| hspb1 | 0.131 | eef1da | -0.098 |
| nop58 | 0.125 | bhmt | -0.096 |
| ncl | 0.125 | zgc:114188 | -0.092 |
| wu:fb97g03 | 0.124 | zgc:158463 | -0.090 |
| fbl | 0.124 | actc1b | -0.087 |
| hmga1a | 0.122 | rps10 | -0.085 |
| npm1a | 0.122 | eif4a1b | -0.085 |
| anp32e | 0.121 | ckmb | -0.084 |
| ilf3b | 0.120 | ndrg2 | -0.083 |
| rbm8a | 0.120 | ckma | -0.083 |
| dkc1 | 0.120 | rps17 | -0.082 |
| syncrip | 0.119 | idh2 | -0.081 |
| lyar | 0.119 | eef1b2 | -0.080 |
| nop56 | 0.118 | atp2a1 | -0.080 |
| srsf1a | 0.117 | pvalb1 | -0.080 |
| hnrnpabb | 0.117 | eno3 | -0.080 |
| hnrnpub | 0.116 | rps29 | -0.079 |
| nucks1a | 0.116 | pvalb2 | -0.078 |
| ptges3b | 0.116 | pabpc4 | -0.078 |
| ppig | 0.115 | mylpfa | -0.077 |
| u2af2b | 0.115 | pmp22a | -0.077 |
| hnrnpaba | 0.114 | mylz3 | -0.076 |
| tbx16 | 0.114 | tpi1b | -0.075 |
| si:ch73-281n10.2 | 0.114 | tnnt3a | -0.075 |
| bms1 | 0.113 | COX3 | -0.074 |
| snrpb | 0.112 | mt-atp6 | -0.074 |
| rpl7l1 | 0.112 | nme2b.1 | -0.074 |
| XLOC-032526 | 0.112 | eno1a | -0.074 |
| nhp2 | 0.111 | rplp2 | -0.073 |
| efnb2b | 0.111 | col1a1a | -0.073 |
| fn1b | 0.110 | col1a2 | -0.073 |
| ddx39ab | 0.110 | ak1 | -0.072 |