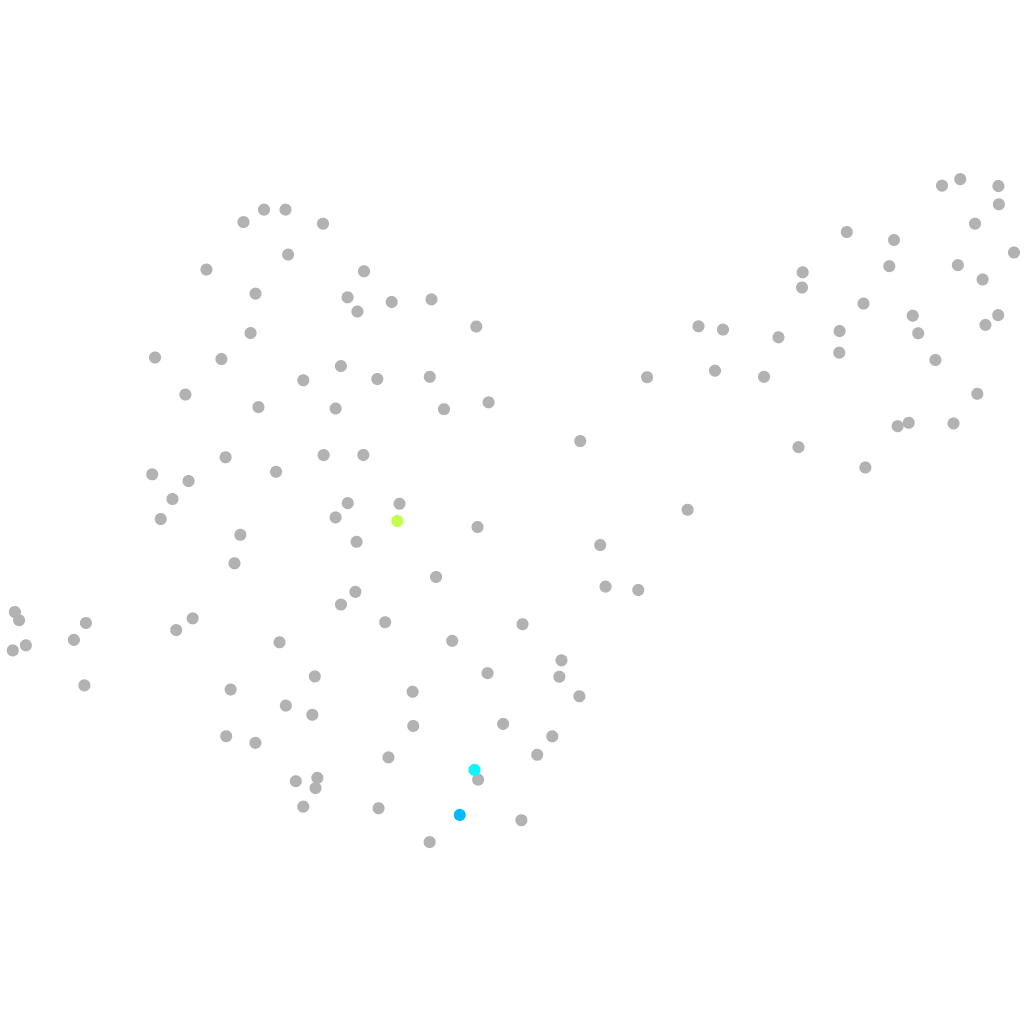
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bbox1 | 0.686 | naca | -0.246 |
| CR627496.1 | 0.686 | rps27.2 | -0.236 |
| CR925797.1 | 0.686 | slc25a5 | -0.227 |
| gna14 | 0.686 | csnk2b | -0.226 |
| si:ch211-139a5.1 | 0.686 | eif3f | -0.223 |
| tbx15 | 0.686 | rpl14 | -0.222 |
| slc28a1 | 0.661 | jpt1b | -0.220 |
| sprn | 0.647 | csnk1a1 | -0.219 |
| sinhcaf | 0.644 | stk35l | -0.218 |
| CR762436.2 | 0.632 | ccng1 | -0.216 |
| srgap1b | 0.624 | ndufc2 | -0.214 |
| myo18ab | 0.583 | c1d | -0.212 |
| unk | 0.581 | ube2e1 | -0.207 |
| zmp:0000000521 | 0.576 | nudc | -0.201 |
| XLOC-006295 | 0.572 | ppt1 | -0.201 |
| lgalsla | 0.572 | si:ch211-276i12.11 | -0.198 |
| LOC108180131 | 0.572 | prkrip1 | -0.197 |
| rx1 | 0.572 | NC-002333.17 | -0.196 |
| XLOC-017482 | 0.572 | rpl38 | -0.192 |
| klhl26 | 0.562 | sept7b | -0.191 |
| cand2 | 0.550 | btf3l4 | -0.191 |
| si:ch73-144d13.7 | 0.549 | fam53b | -0.190 |
| trim45 | 0.548 | rps15 | -0.189 |
| LOC100150038 | 0.545 | srsf1a | -0.188 |
| acer2 | 0.535 | eif1axb | -0.188 |
| slc10a3 | 0.530 | sumo1 | -0.186 |
| parpbp | 0.524 | atp5pd | -0.186 |
| CR936462.1 | 0.524 | rpl7a | -0.185 |
| tmem145 | 0.523 | ccdc25 | -0.183 |
| pick1 | 0.522 | hsp90ab1 | -0.182 |
| LOC108179808 | 0.519 | cct3 | -0.181 |
| ppp4r1l | 0.519 | mt-nd4 | -0.179 |
| zdhhc14 | 0.514 | eif3ba | -0.178 |
| si:dkey-181m9.8 | 0.512 | pa2g4b | -0.178 |
| myo1ca | 0.506 | nap1l1 | -0.177 |