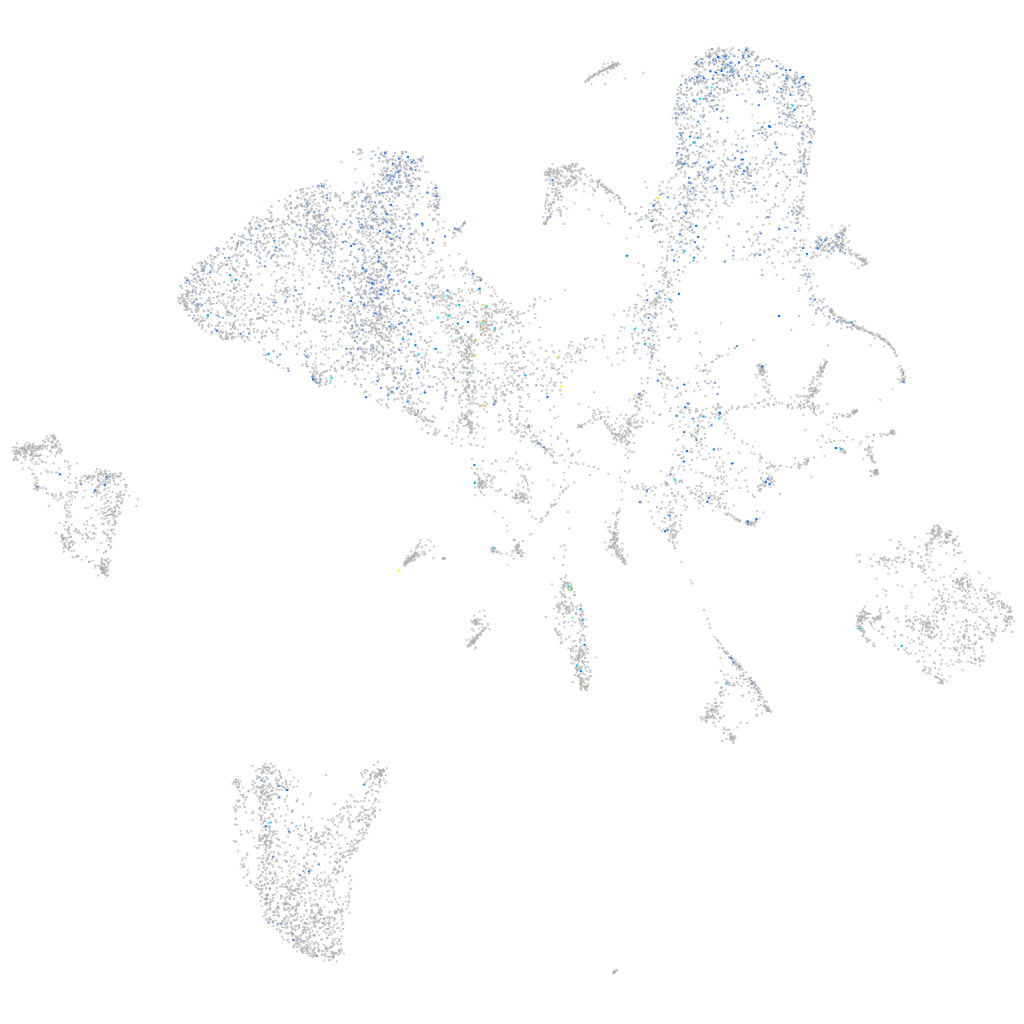
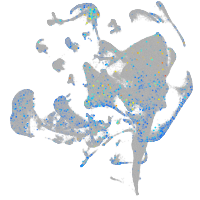
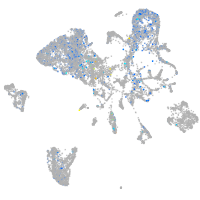
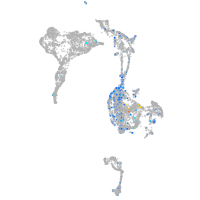
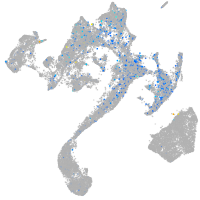
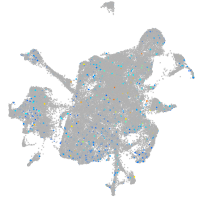
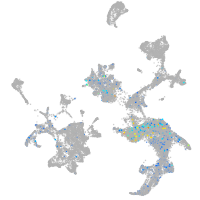
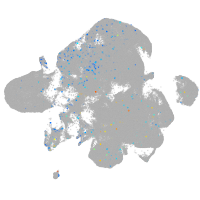
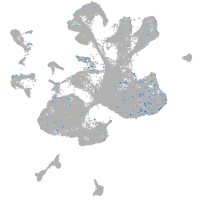
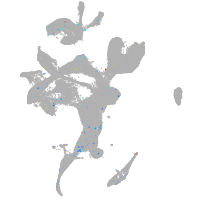
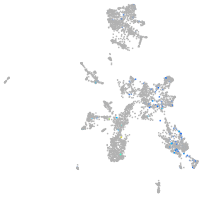
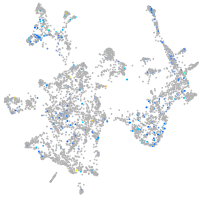
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoc2 | 0.162 | si:ch211-195b11.3 | -0.080 |
| sult2st2 | 0.161 | npdc1a | -0.079 |
| gcshb | 0.158 | slc38a5b | -0.077 |
| gstr | 0.154 | marcksb | -0.077 |
| gstt1a | 0.154 | stard10 | -0.075 |
| suclg1 | 0.153 | ctnnb1 | -0.074 |
| apoa4b.1 | 0.153 | rtn1a | -0.074 |
| mdh1aa | 0.151 | hnrnpa1a | -0.071 |
| prdx2 | 0.150 | hmgb3a | -0.071 |
| eno3 | 0.149 | anxa5b | -0.070 |
| cdo1 | 0.148 | hmgb1b | -0.070 |
| glud1b | 0.147 | anp32e | -0.070 |
| scp2a | 0.147 | icn | -0.070 |
| rbp2a | 0.146 | ctrl | -0.070 |
| sdr16c5b | 0.146 | ywhaz | -0.069 |
| dhrs9 | 0.146 | cfl1l | -0.069 |
| gpx4a | 0.145 | epcam | -0.069 |
| rdh1 | 0.145 | akap12b | -0.069 |
| fdx1 | 0.144 | zgc:92380 | -0.068 |
| fbp1b | 0.143 | marcksl1b | -0.068 |
| apobb.1 | 0.142 | prss59.2 | -0.068 |
| agmo | 0.142 | hmgb1a | -0.067 |
| gapdh | 0.142 | ier2b | -0.067 |
| afp4 | 0.141 | CELA1 (1 of many) | -0.067 |
| aldh9a1a.1 | 0.137 | zgc:112160 | -0.067 |
| ugt1a7 | 0.137 | zgc:136461 | -0.067 |
| cyp8b1 | 0.136 | chd4b | -0.067 |
| pla2g12b | 0.135 | ctrb1 | -0.067 |
| fabp2 | 0.135 | pnrc2 | -0.066 |
| tpi1b | 0.135 | ela3l | -0.066 |
| msrb2 | 0.134 | prss59.1 | -0.066 |
| sult3st3 | 0.134 | hnrnpa0a | -0.066 |
| sod1 | 0.134 | ela2 | -0.066 |
| rmdn1 | 0.134 | cpb1 | -0.065 |
| sult3st2 | 0.133 | gapdhs | -0.065 |