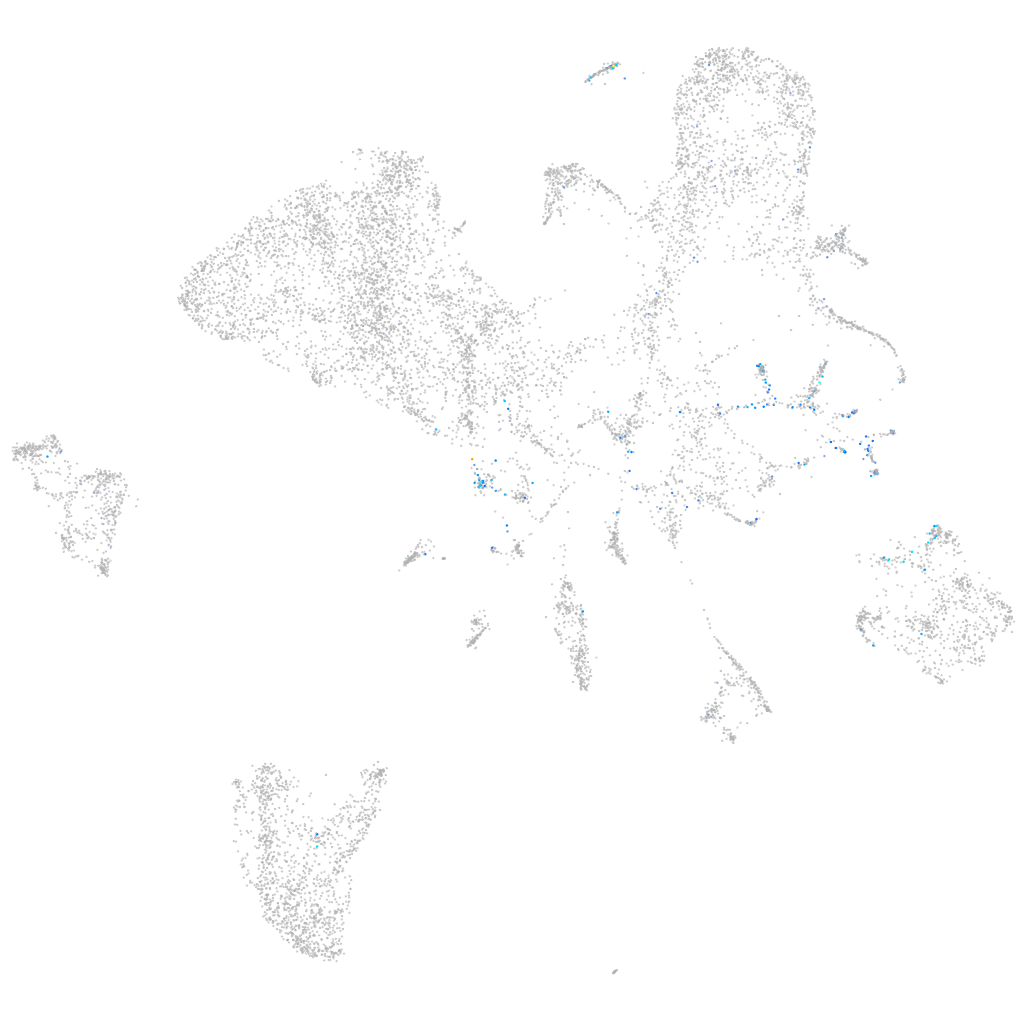
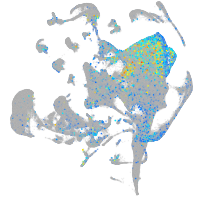
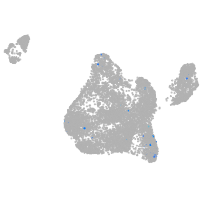
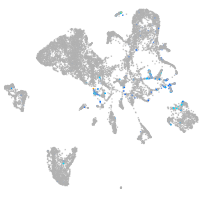
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.262 | ahcy | -0.118 |
| neurod1 | 0.238 | aldob | -0.117 |
| id4 | 0.237 | gapdh | -0.114 |
| pcsk1nl | 0.236 | gamt | -0.104 |
| mir7a-1 | 0.236 | gatm | -0.097 |
| syt1a | 0.233 | fbp1b | -0.093 |
| vamp2 | 0.229 | aldh6a1 | -0.092 |
| insm1b | 0.225 | nupr1b | -0.091 |
| pax6b | 0.223 | mat1a | -0.088 |
| atpv0e2 | 0.222 | bhmt | -0.088 |
| insm1a | 0.220 | gstr | -0.088 |
| LOC100537384 | 0.217 | eno3 | -0.086 |
| myt1b | 0.217 | cx32.3 | -0.086 |
| si:dkey-153k10.9 | 0.214 | apoa4b.1 | -0.085 |
| atp1a3a | 0.212 | gnmt | -0.083 |
| vat1 | 0.207 | aqp12 | -0.082 |
| amph | 0.203 | aldh7a1 | -0.082 |
| pcsk1 | 0.202 | agxtb | -0.082 |
| map1b | 0.201 | apoa1b | -0.081 |
| tmsb | 0.200 | glud1b | -0.080 |
| aldocb | 0.199 | scp2a | -0.080 |
| scg2a | 0.199 | apoc2 | -0.079 |
| atp6v0cb | 0.197 | gstp1 | -0.079 |
| zgc:101731 | 0.196 | acadm | -0.079 |
| rnasekb | 0.194 | gstt1a | -0.078 |
| nkx2.2a | 0.193 | agxta | -0.077 |
| gpc1a | 0.192 | mgst1.2 | -0.077 |
| syt11a | 0.192 | cebpd | -0.077 |
| fcer1g | 0.191 | shmt1 | -0.076 |
| rtn1b | 0.189 | nipsnap3a | -0.076 |
| tusc3 | 0.188 | tktb | -0.076 |
| tuba1c | 0.187 | hdlbpa | -0.076 |
| tmem178b | 0.187 | pnp4b | -0.076 |
| zgc:65894 | 0.187 | pgm1 | -0.075 |
| gap43 | 0.186 | eef1da | -0.075 |