wu:fi46h04
ZFIN 
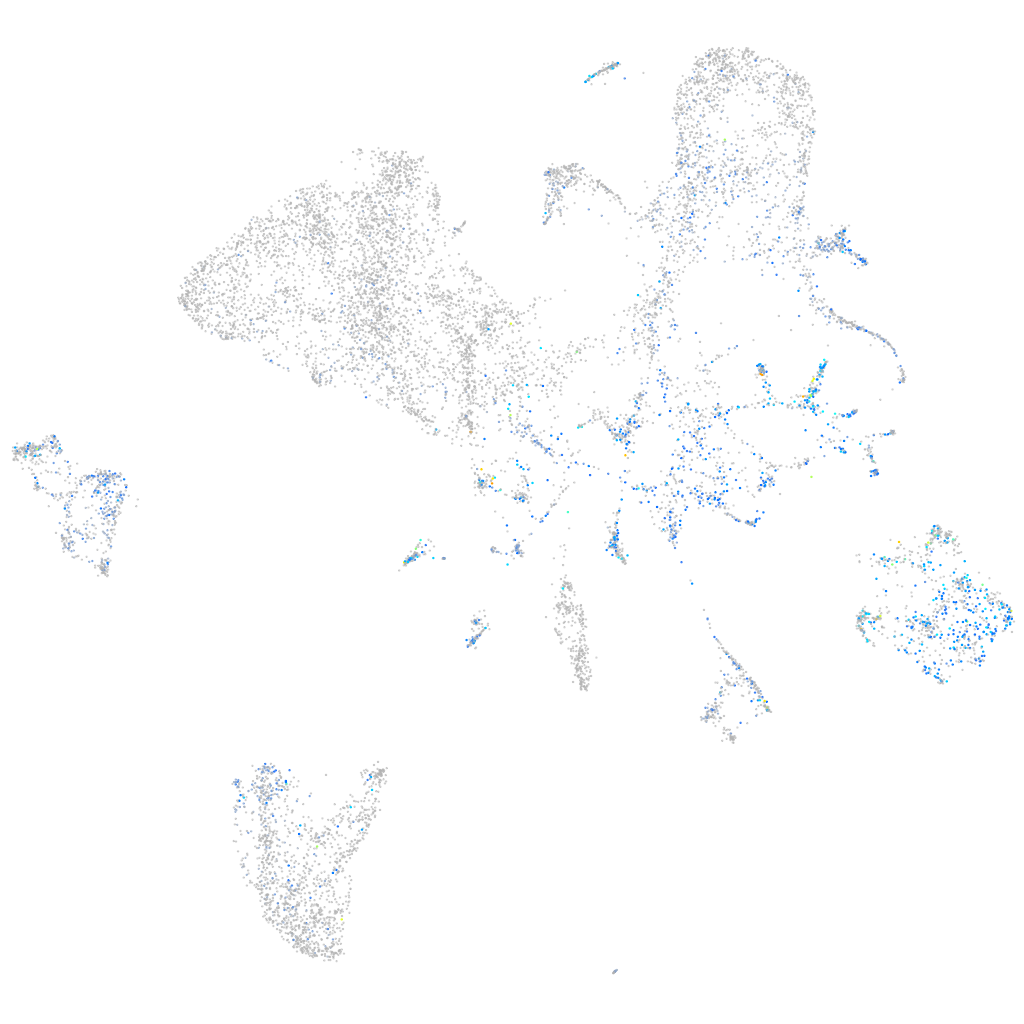
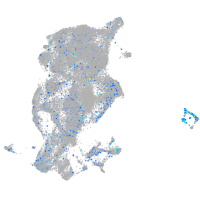
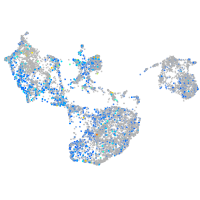
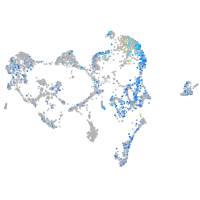
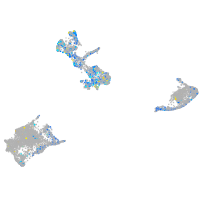
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.288 | gamt | -0.281 |
| hmgn6 | 0.287 | gapdh | -0.274 |
| khdrbs1a | 0.280 | ahcy | -0.270 |
| syncrip | 0.278 | gatm | -0.261 |
| marcksl1b | 0.277 | apoa1b | -0.247 |
| nucks1a | 0.273 | bhmt | -0.239 |
| si:ch211-222l21.1 | 0.272 | apoa4b.1 | -0.238 |
| si:ch73-1a9.3 | 0.271 | fbp1b | -0.235 |
| h3f3d | 0.269 | mat1a | -0.234 |
| marcksb | 0.269 | afp4 | -0.234 |
| hnrnpa0a | 0.268 | apoc2 | -0.233 |
| chd7 | 0.266 | gpx4a | -0.225 |
| acin1a | 0.263 | apoa2 | -0.224 |
| hnrnpa0b | 0.262 | eno3 | -0.221 |
| anp32a | 0.258 | scp2a | -0.221 |
| smarca4a | 0.256 | glud1b | -0.218 |
| hmgb3a | 0.256 | agxtb | -0.218 |
| hmga1a | 0.255 | suclg1 | -0.215 |
| cirbpa | 0.255 | grhprb | -0.211 |
| hdac1 | 0.253 | aldob | -0.211 |
| gnb1b | 0.250 | pnp4b | -0.210 |
| si:ch73-281n10.2 | 0.248 | apobb.1 | -0.209 |
| hmgb2a | 0.248 | mgst1.2 | -0.208 |
| ptmab | 0.248 | apom | -0.208 |
| cirbpb | 0.248 | tfa | -0.207 |
| hnrnpabb | 0.248 | fabp10a | -0.205 |
| seta | 0.247 | gstt1a | -0.205 |
| hnrnpa1b | 0.247 | cx32.3 | -0.205 |
| hp1bp3 | 0.246 | serpina1 | -0.205 |
| h2afvb | 0.246 | abat | -0.204 |
| hmgn7 | 0.246 | serpina1l | -0.203 |
| akap12b | 0.245 | ces2 | -0.203 |
| rhoaa | 0.243 | zgc:123103 | -0.203 |
| hmgb1a | 0.241 | rbp2b | -0.202 |
| atrx | 0.240 | rbp4 | -0.200 |