wu:fb16f03
ZFIN 
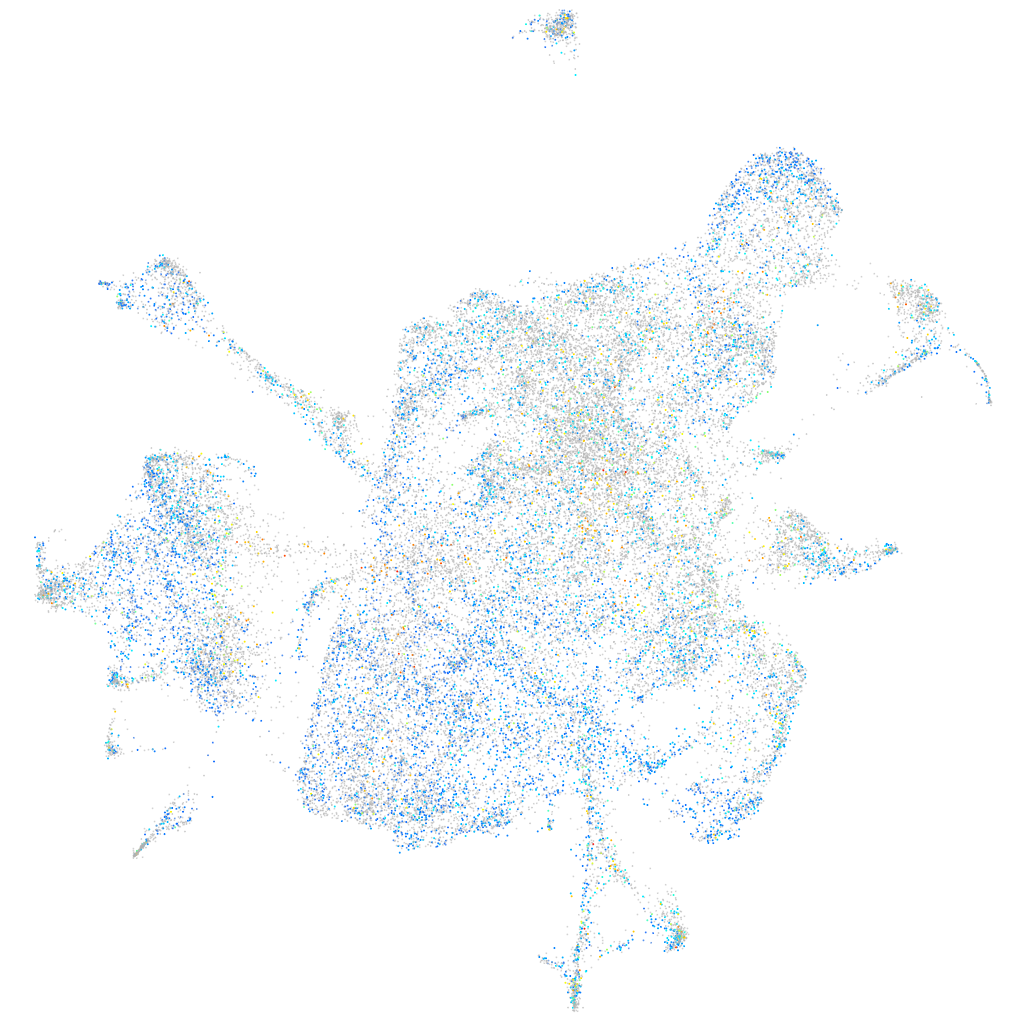
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mt-nd4 | 0.084 | pvalb2 | -0.064 |
| NC-002333.4 | 0.083 | pvalb1 | -0.061 |
| mt-cyb | 0.080 | apoa2 | -0.054 |
| canx | 0.078 | rplp1 | -0.053 |
| atp1a1a.1 | 0.076 | actc1b | -0.053 |
| igf2bp1 | 0.076 | apoa1b | -0.051 |
| mt-nd2 | 0.074 | rbp4 | -0.048 |
| eif4g1a | 0.072 | faua | -0.048 |
| pfn2l | 0.072 | mylz3 | -0.044 |
| mt-co1 | 0.071 | rpl39 | -0.044 |
| asph | 0.069 | rps25 | -0.043 |
| cirbpa | 0.068 | prss59.2 | -0.043 |
| mt-atp6 | 0.068 | dap1b | -0.043 |
| hnrnpa1a | 0.067 | rps14 | -0.042 |
| fkbp9 | 0.067 | rpl10a | -0.041 |
| hnrnpa0a | 0.066 | rpl27a | -0.041 |
| aldoaa | 0.066 | rps23 | -0.041 |
| pbx4 | 0.065 | rpl19 | -0.041 |
| akap12b | 0.065 | CABZ01092746.1 | -0.041 |
| eif5a2 | 0.065 | rps15a | -0.041 |
| ptges3b | 0.065 | rpl10 | -0.040 |
| ctnnb1 | 0.065 | ifitm1 | -0.040 |
| cfl1 | 0.064 | naca | -0.039 |
| smarca4a | 0.063 | rps26l | -0.038 |
| mt-nd1 | 0.062 | rpl37 | -0.038 |
| si:dkeyp-106c3.1 | 0.062 | eef1da | -0.038 |
| seta | 0.062 | prss59.1 | -0.038 |
| cdh2 | 0.062 | rps19 | -0.037 |
| rpn1 | 0.062 | rps27.1 | -0.037 |
| mt-nd3 | 0.062 | rps7 | -0.036 |
| acin1a | 0.061 | CELA1 (1 of many) | -0.036 |
| CABZ01072614.1 | 0.061 | rpl23 | -0.036 |
| eif4a1a | 0.061 | rps21 | -0.036 |
| chd7 | 0.060 | ecrg4a | -0.036 |
| prmt1 | 0.060 | rpl22 | -0.036 |