"wingless-type MMTV integration site family, member 9A"
ZFIN 
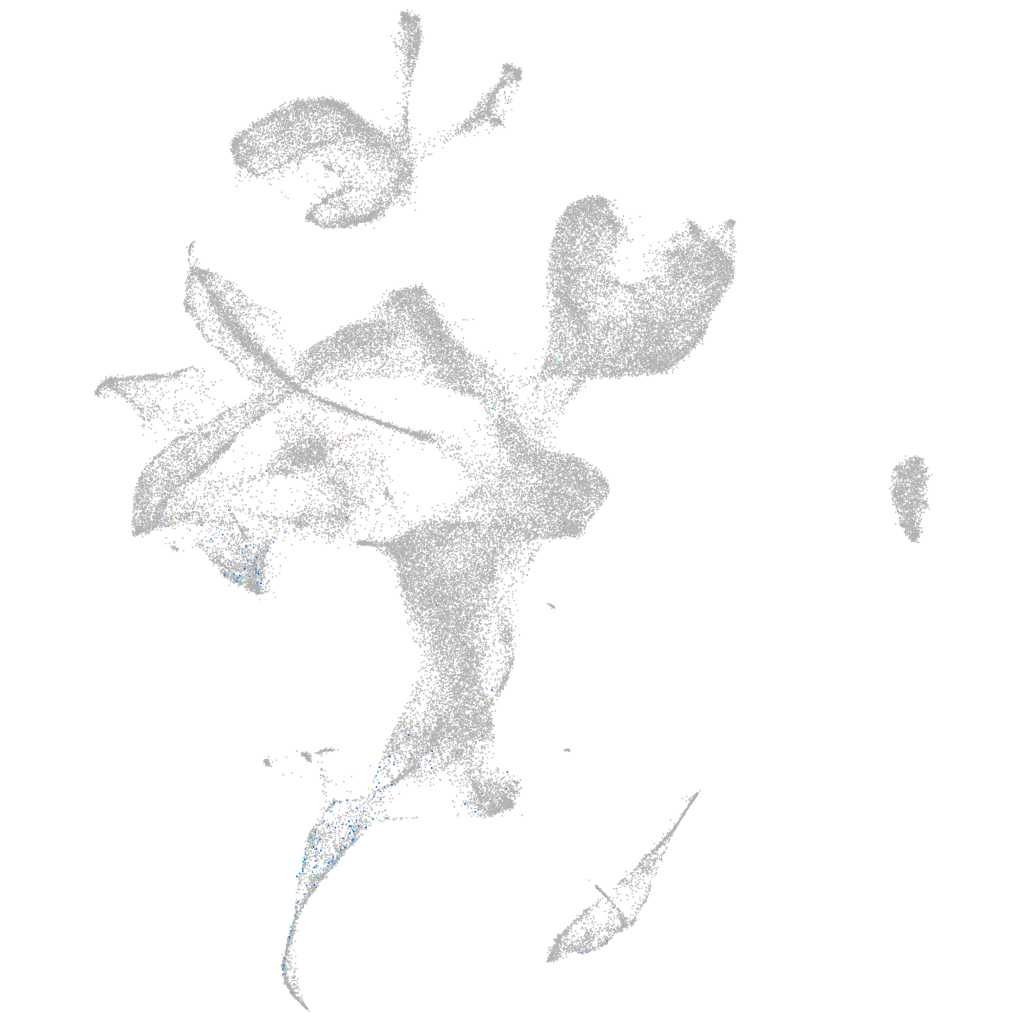
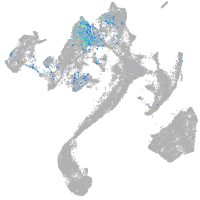
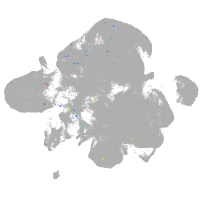
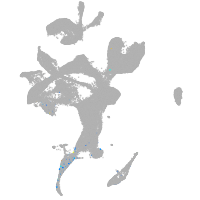
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rab38 | 0.138 | ptmab | -0.062 |
| slc45a2 | 0.137 | si:ch73-1a9.3 | -0.046 |
| pmela | 0.133 | gpm6aa | -0.043 |
| dct | 0.133 | hmgn6 | -0.041 |
| gpr143 | 0.131 | si:ch211-137a8.4 | -0.037 |
| sparc | 0.129 | tuba1c | -0.036 |
| tyrp1b | 0.128 | ptmaa | -0.036 |
| tyrp1a | 0.126 | tuba1a | -0.036 |
| pmelb | 0.126 | fabp7a | -0.034 |
| tspan36 | 0.125 | cadm3 | -0.033 |
| oca2 | 0.124 | rorb | -0.031 |
| LOC103910009 | 0.123 | hmgb1b | -0.031 |
| col4a6 | 0.123 | h2afva | -0.030 |
| col4a5 | 0.121 | hmgb1a | -0.030 |
| pah | 0.121 | hmgn2 | -0.029 |
| fstl1b | 0.120 | sumo3a | -0.029 |
| sytl2b | 0.120 | hnrnpaba | -0.029 |
| CT025909.2 | 0.118 | gpm6ab | -0.027 |
| qdpra | 0.118 | foxg1b | -0.027 |
| agtrap | 0.117 | marcksl1b | -0.026 |
| tyr | 0.116 | anp32e | -0.025 |
| tspan10 | 0.113 | cirbpb | -0.025 |
| zgc:110591 | 0.113 | neurod4 | -0.025 |
| gstp1 | 0.113 | nova2 | -0.025 |
| fbn2b | 0.111 | gapdhs | -0.024 |
| wnt2 | 0.111 | ckbb | -0.024 |
| slc7a9 | 0.111 | marcksb | -0.023 |
| rab27a | 0.110 | rtn1a | -0.023 |
| abcg2d | 0.109 | h3f3d | -0.023 |
| slc24a5 | 0.108 | hes6 | -0.023 |
| col9a2 | 0.108 | nono | -0.023 |
| ldlrap1a | 0.107 | hnrnpa0a | -0.023 |
| cracr2ab | 0.107 | mdkb | -0.022 |
| pttg1ipb | 0.105 | si:ch1073-429i10.3.1 | -0.022 |
| bace2 | 0.105 | khdrbs1a | -0.022 |