"wingless-type MMTV integration site family, member 2Ba"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis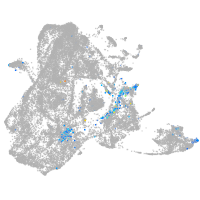 |
periderm |
fin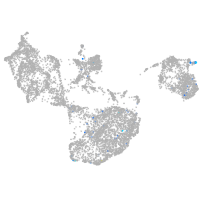 |
ionocytes / mucous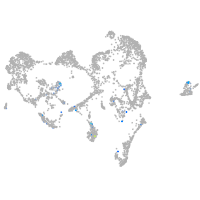 |
pigment cells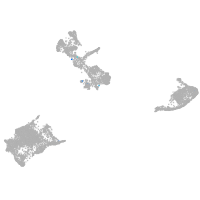 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| desmb | 0.254 | ednraa | -0.088 |
| kcnk18 | 0.247 | emid1 | -0.084 |
| cald1b | 0.240 | si:dkey-261h17.1 | -0.075 |
| cabp1a | 0.238 | h3f3d | -0.070 |
| tpm2 | 0.234 | prss35 | -0.067 |
| csrp1b | 0.225 | NC-002333.4 | -0.067 |
| myl6 | 0.221 | rplp2l | -0.066 |
| mylkb | 0.214 | efnb1 | -0.066 |
| actc1a | 0.212 | notch3 | -0.066 |
| si:ch211-137i24.10 | 0.212 | mmp2 | -0.065 |
| acta2 | 0.203 | hmga1a | -0.065 |
| pnp4a | 0.202 | mfap5 | -0.065 |
| lxn | 0.199 | col12a1a | -0.065 |
| pdlim3a | 0.194 | si:ch73-1a9.3 | -0.063 |
| myh11a | 0.190 | rasl12 | -0.062 |
| cnn1b | 0.190 | sparc | -0.061 |
| tagln | 0.189 | palm1b | -0.061 |
| gem | 0.186 | cdc42ep4a | -0.061 |
| ckba | 0.186 | mef2cb | -0.061 |
| smtna | 0.185 | nme2b.1 | -0.060 |
| myl9a | 0.185 | si:ch73-281n10.2 | -0.060 |
| acta1b | 0.184 | rbpms2b | -0.059 |
| tpm1 | 0.184 | top1l | -0.059 |
| fbxl22 | 0.184 | alcama | -0.059 |
| nog1 | 0.180 | hif1al | -0.058 |
| ckbb | 0.179 | cd248a | -0.058 |
| fhl1a | 0.179 | vim | -0.057 |
| si:ch211-62a1.3 | 0.175 | foxc1b | -0.057 |
| iscub | 0.171 | tenm3 | -0.057 |
| vcla | 0.171 | cxcl12b | -0.056 |
| ppp1cbl | 0.170 | s1pr3a | -0.056 |
| pgm5 | 0.163 | ctsk | -0.056 |
| acta1a | 0.161 | tgfb2 | -0.056 |
| XLOC-041870 | 0.160 | ackr3b | -0.055 |
| zgc:110699 | 0.159 | fbln5 | -0.055 |




