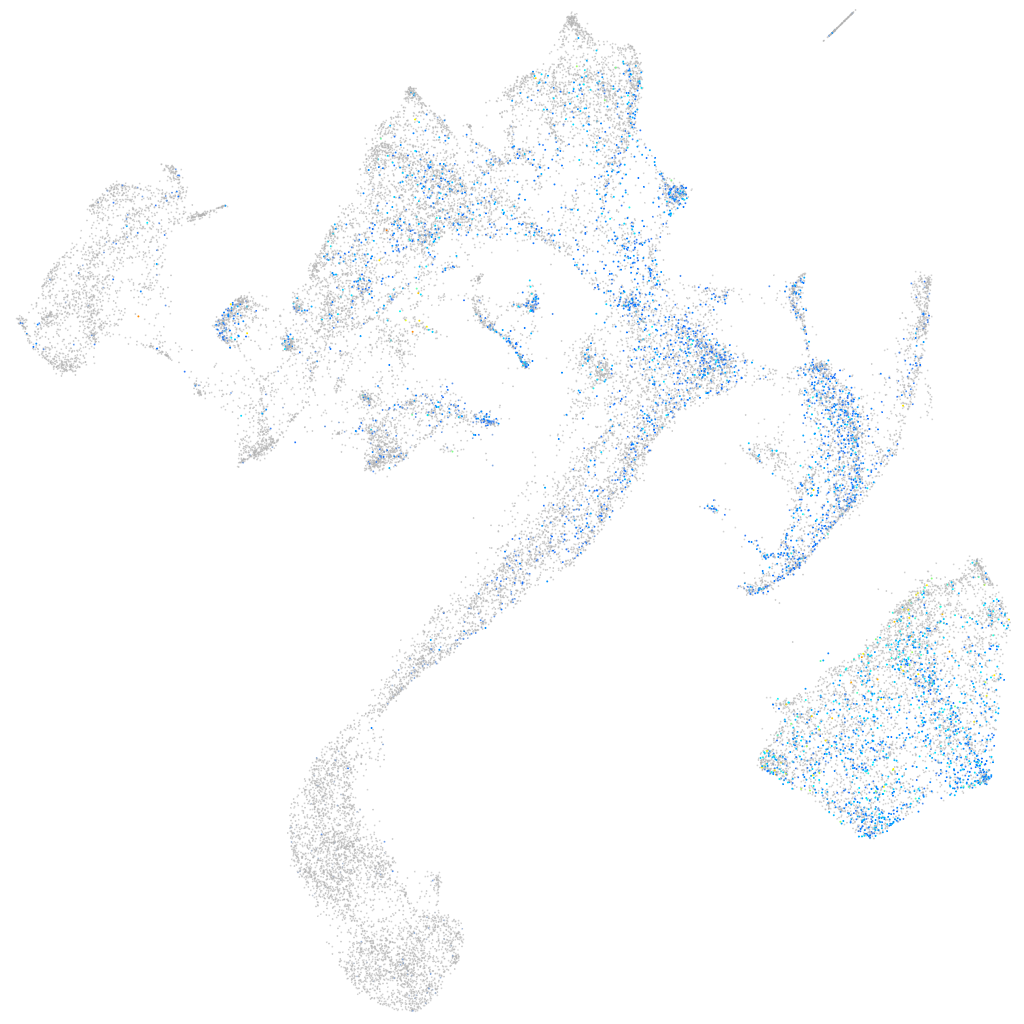
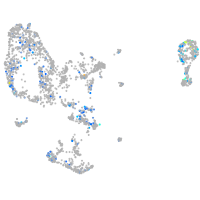
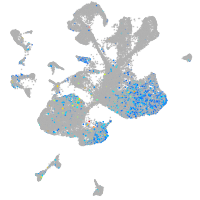
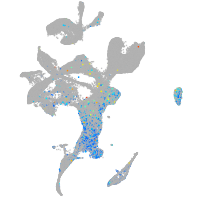
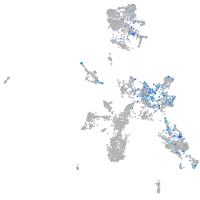
WEE1 G2 checkpoint kinase
ZFIN 
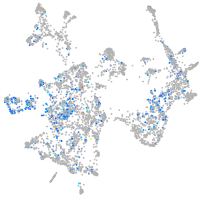
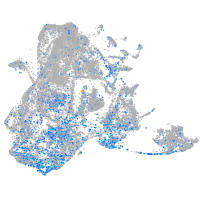
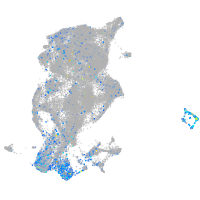
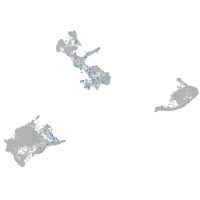
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.276 | actc1b | -0.225 |
| hmgb2a | 0.263 | ak1 | -0.197 |
| tpx2 | 0.256 | ckma | -0.197 |
| anp32b | 0.255 | ckmb | -0.194 |
| ccnb1 | 0.253 | ttn.2 | -0.194 |
| seta | 0.251 | aldoab | -0.193 |
| hmga1a | 0.251 | atp2a1 | -0.193 |
| cx43.4 | 0.247 | tmem38a | -0.187 |
| anp32a | 0.246 | tnnc2 | -0.187 |
| cbx3a | 0.246 | ttn.1 | -0.184 |
| si:ch211-222l21.1 | 0.243 | acta1b | -0.182 |
| ube2c | 0.241 | neb | -0.180 |
| mad2l1 | 0.240 | mylpfa | -0.180 |
| cdca8 | 0.239 | tpma | -0.179 |
| hmgb2b | 0.239 | pabpc4 | -0.177 |
| hnrnpa0b | 0.238 | mybphb | -0.176 |
| tuba8l4 | 0.237 | desma | -0.176 |
| khdrbs1a | 0.235 | ldb3a | -0.173 |
| hnrnpabb | 0.235 | ldb3b | -0.172 |
| ppm1g | 0.234 | srl | -0.171 |
| plk1 | 0.233 | nme2b.2 | -0.171 |
| cirbpa | 0.232 | cav3 | -0.170 |
| nusap1 | 0.232 | gamt | -0.170 |
| hnrnpa1b | 0.231 | eno1a | -0.169 |
| si:ch73-281n10.2 | 0.231 | CABZ01078594.1 | -0.169 |
| si:ch211-288g17.3 | 0.230 | si:ch73-367p23.2 | -0.168 |
| hnrnpaba | 0.230 | actn3b | -0.167 |
| syncrip | 0.229 | mylz3 | -0.167 |
| marcksb | 0.228 | eno3 | -0.167 |
| banf1 | 0.228 | acta1a | -0.167 |
| ptmab | 0.227 | tnnt3a | -0.167 |
| nucks1a | 0.227 | gapdh | -0.166 |
| ccna2 | 0.226 | actn3a | -0.166 |
| hnrnpub | 0.225 | rpl37 | -0.164 |
| si:ch73-1a9.3 | 0.224 | atp5if1b | -0.164 |