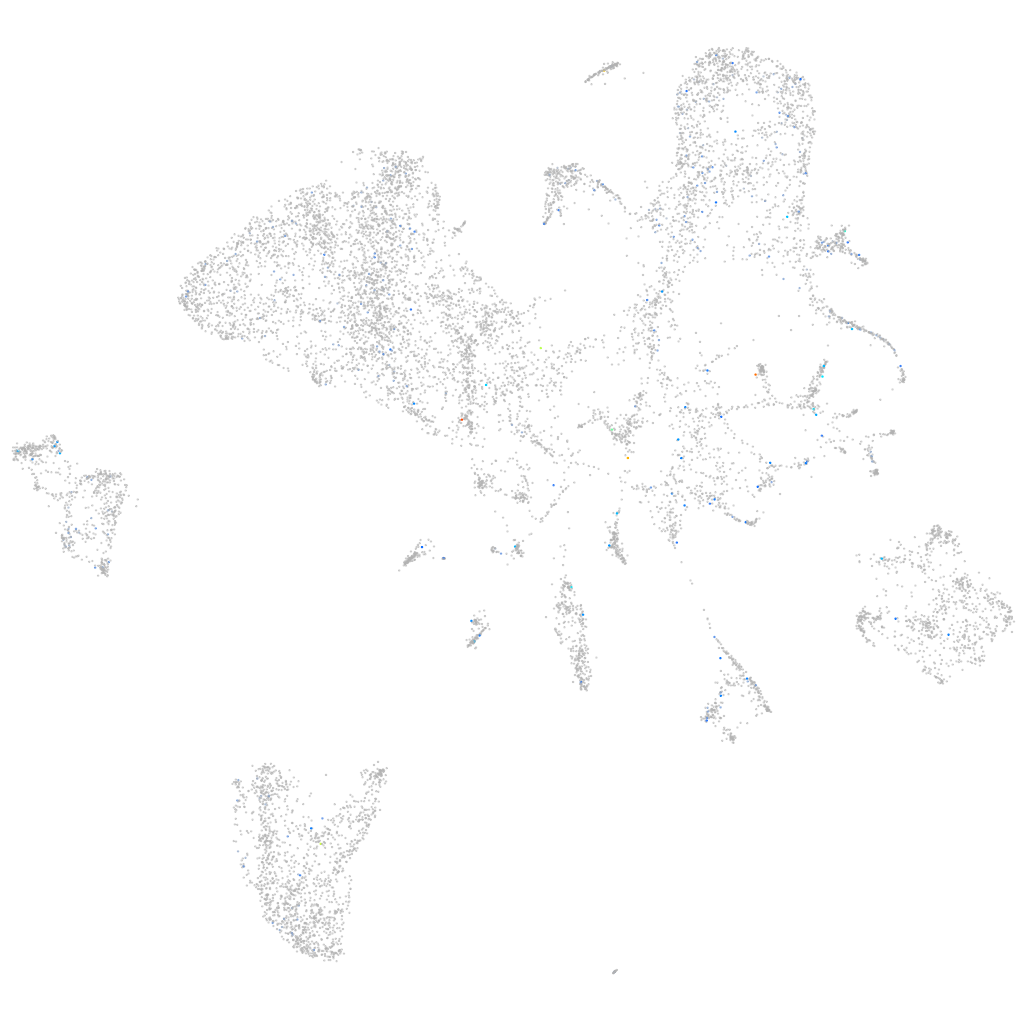
WD repeat domain 48a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01072563.1 | 0.298 | bhmt | -0.051 |
| LOC110438234 | 0.260 | gamt | -0.046 |
| LOC103910960 | 0.238 | apoc1 | -0.045 |
| calcoco1b | 0.220 | aqp12 | -0.044 |
| si:ch211-219a15.3 | 0.216 | agxtb | -0.042 |
| gcm2 | 0.210 | ttc36 | -0.041 |
| bgnb | 0.177 | hao1 | -0.040 |
| ms4a17a.7 | 0.172 | fabp10a | -0.040 |
| otogl | 0.171 | grhprb | -0.039 |
| edil3b | 0.169 | uox | -0.039 |
| BX950188.1 | 0.167 | agxta | -0.039 |
| csrp1b | 0.164 | ttr | -0.039 |
| XLOC-003138 | 0.164 | si:dkey-86l18.10 | -0.039 |
| LOC103910907 | 0.163 | zgc:92744 | -0.039 |
| LOC103911600 | 0.163 | rps12 | -0.038 |
| CU019663.1 | 0.158 | gc | -0.038 |
| klf1 | 0.156 | apom | -0.038 |
| si:dkey-193b15.8 | 0.155 | tfa | -0.038 |
| calhm2 | 0.151 | gatm | -0.038 |
| LOC100536746 | 0.148 | zgc:123103 | -0.038 |
| LOC100331291 | 0.147 | ces2 | -0.038 |
| si:ch73-71d17.2 | 0.137 | serpina1l | -0.038 |
| XLOC-036961 | 0.135 | rbp2b | -0.038 |
| esyt3 | 0.133 | apoa1b | -0.037 |
| XLOC-026937 | 0.130 | serpina1 | -0.037 |
| LO017816.1 | 0.129 | gpd1b | -0.037 |
| si:ch211-51i16.1 | 0.128 | nupr1b | -0.037 |
| cacna2d2a | 0.128 | fetub | -0.036 |
| LOC110440222 | 0.128 | cpn1 | -0.036 |
| bsnb | 0.128 | kng1 | -0.036 |
| crhr1 | 0.128 | gapdh | -0.036 |
| XLOC-026841 | 0.126 | ahcy | -0.036 |
| LOC110439953 | 0.125 | apoa2 | -0.036 |
| LOC110438401 | 0.123 | ambp | -0.035 |
| nhsb | 0.118 | fgg | -0.035 |