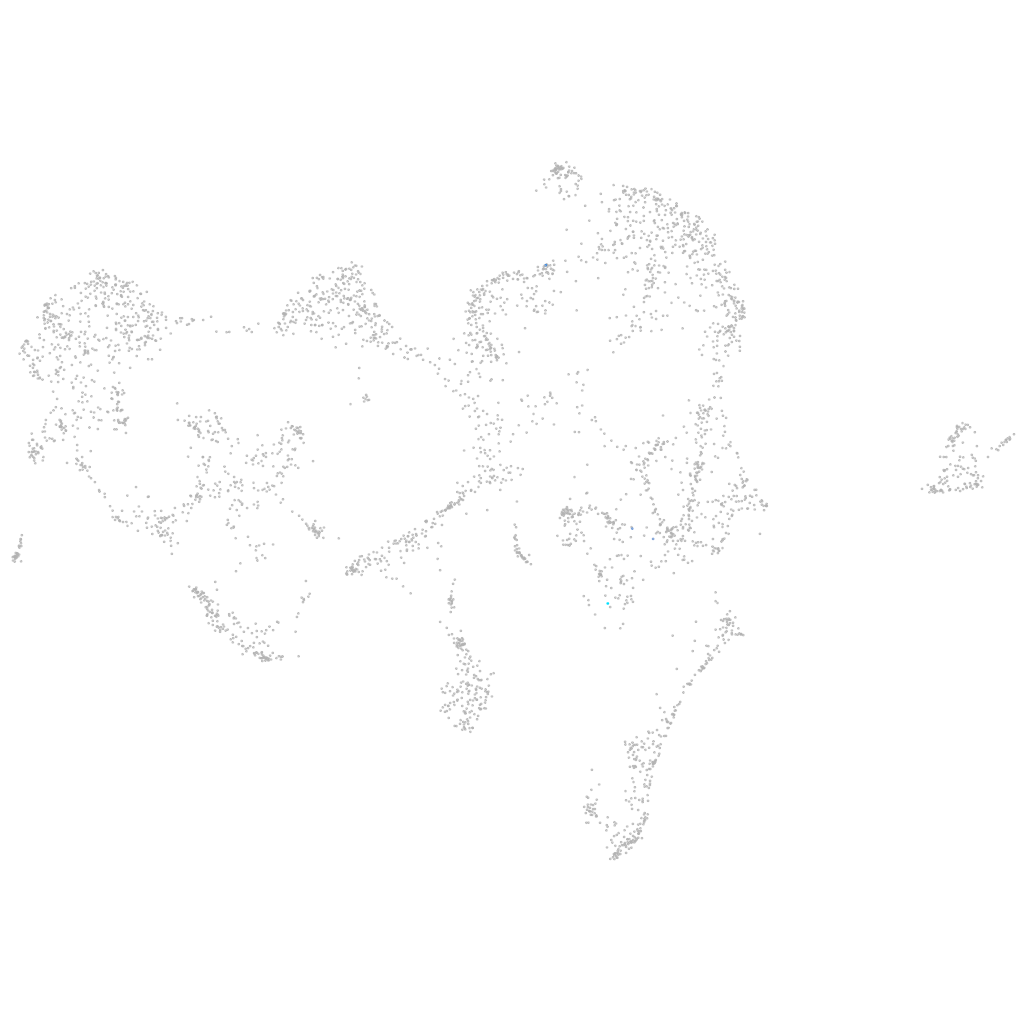
WD repeat domain 27
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccdc175 | 0.853 | ier2a | -0.035 |
| LOC100536937 | 0.853 | ier5 | -0.034 |
| znf1166 | 0.853 | aldob | -0.034 |
| CU550719.2 | 0.853 | chac1 | -0.033 |
| si:dkey-222b8.4 | 0.850 | tob1b | -0.033 |
| dnaaf3l | 0.811 | zgc:162730 | -0.032 |
| lrguk | 0.790 | cldn7b | -0.031 |
| CU633857.1 | 0.754 | si:dkey-87o1.2 | -0.030 |
| si:ch1073-181h11.2 | 0.711 | phlda2 | -0.030 |
| si:dkeyp-86c4.1 | 0.700 | ssr3 | -0.030 |
| rsph3 | 0.689 | junba | -0.030 |
| cfap300 | 0.685 | tmed7 | -0.029 |
| cfap100 | 0.684 | atf4a | -0.029 |
| c18h15orf65 | 0.647 | cnbpb | -0.029 |
| got1l1 | 0.632 | jun | -0.029 |
| drc1 | 0.629 | oclna | -0.029 |
| spata18 | 0.619 | ddit3 | -0.029 |
| CR855277.2 | 0.603 | ssr2 | -0.029 |
| mustn1a | 0.601 | selenow1 | -0.028 |
| lrrc51 | 0.599 | eif2s1b | -0.028 |
| dnali1 | 0.552 | lgals3b | -0.028 |
| tbata | 0.538 | gmds | -0.027 |
| znf1016 | 0.535 | chmp1b | -0.027 |
| ccdc146 | 0.531 | fuca1.1 | -0.027 |
| ccdc13 | 0.524 | cyt1 | -0.027 |
| si:dkeyp-69c1.9 | 0.519 | pdia3 | -0.027 |
| nme5 | 0.502 | pycard | -0.027 |
| katnal2 | 0.486 | s100v2 | -0.026 |
| tekt1 | 0.479 | cebpb | -0.025 |
| foxj1b | 0.473 | ccni | -0.025 |
| dnaaf4 | 0.472 | calm3b | -0.025 |
| si:dkey-163f12.6 | 0.467 | srsf5a | -0.025 |
| cd37 | 0.459 | gdi2 | -0.025 |
| akna | 0.449 | anxa4 | -0.025 |
| mcoln2 | 0.448 | psma1 | -0.025 |