WD repeat and HMG-box DNA binding protein 1
ZFIN 
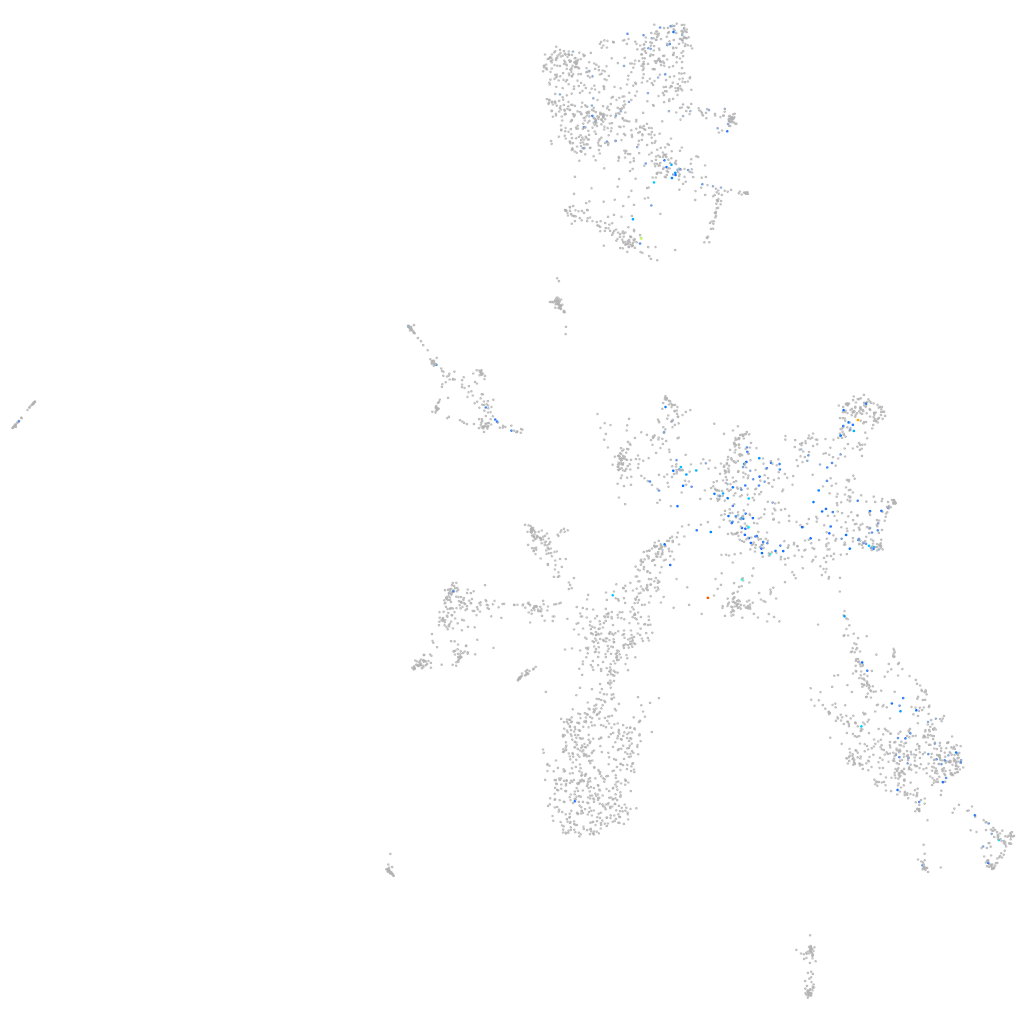
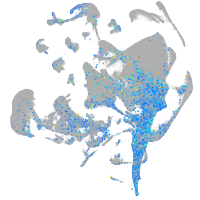
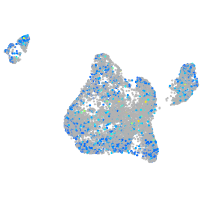
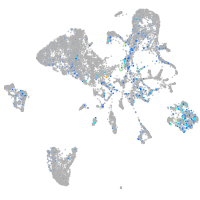
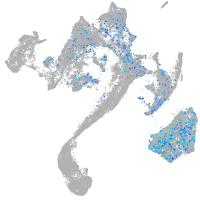
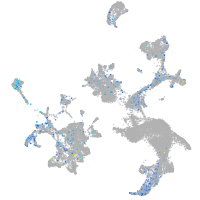
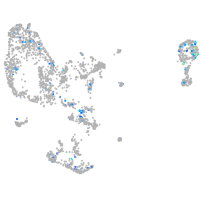
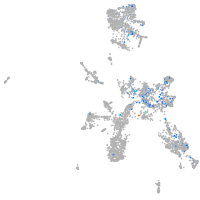
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gins2 | 0.356 | txn | -0.146 |
| pcna | 0.332 | cldnh | -0.137 |
| mcm7 | 0.325 | CR383676.1 | -0.129 |
| rpa3 | 0.315 | anxa5b | -0.122 |
| zgc:110540 | 0.309 | gapdhs | -0.121 |
| CABZ01005379.1 | 0.307 | rnasekb | -0.120 |
| esco2 | 0.304 | tmem59 | -0.119 |
| slbp | 0.302 | rtn1a | -0.117 |
| rrm2 | 0.301 | vamp2 | -0.116 |
| nasp | 0.299 | fxyd1 | -0.115 |
| dut | 0.298 | gabarapa | -0.112 |
| rrm1 | 0.297 | gnb1a | -0.112 |
| hells | 0.296 | selenow2b | -0.112 |
| rpa2 | 0.295 | atp6v0cb | -0.111 |
| asf1ba | 0.294 | bik | -0.109 |
| fen1 | 0.287 | pvalb5 | -0.109 |
| prim2 | 0.284 | calm1a | -0.108 |
| cdc6 | 0.279 | gstp1 | -0.108 |
| chtf8 | 0.279 | tspan2a | -0.107 |
| stmn1a | 0.272 | selenow1 | -0.107 |
| tyms | 0.271 | wu:fb18f06 | -0.105 |
| msh6 | 0.270 | gabarapb | -0.105 |
| chaf1a | 0.270 | ccni | -0.104 |
| lig1 | 0.270 | si:dkeyp-75h12.5 | -0.104 |
| mcm4 | 0.269 | zgc:65894 | -0.103 |
| tk1 | 0.269 | nfe2l2a | -0.103 |
| dnmt1 | 0.268 | atp6v1g1 | -0.103 |
| mcm2 | 0.267 | h1f0 | -0.102 |
| mcm5 | 0.267 | nupr1a | -0.102 |
| orc4 | 0.265 | adcyap1b | -0.102 |
| mcm6 | 0.263 | scg2a | -0.101 |
| rfc4 | 0.256 | selenot1a | -0.101 |
| mybl2b | 0.255 | atp6v1e1b | -0.100 |
| dnajc9 | 0.255 | chga | -0.099 |
| rnaseh2a | 0.250 | camk2n1a | -0.098 |