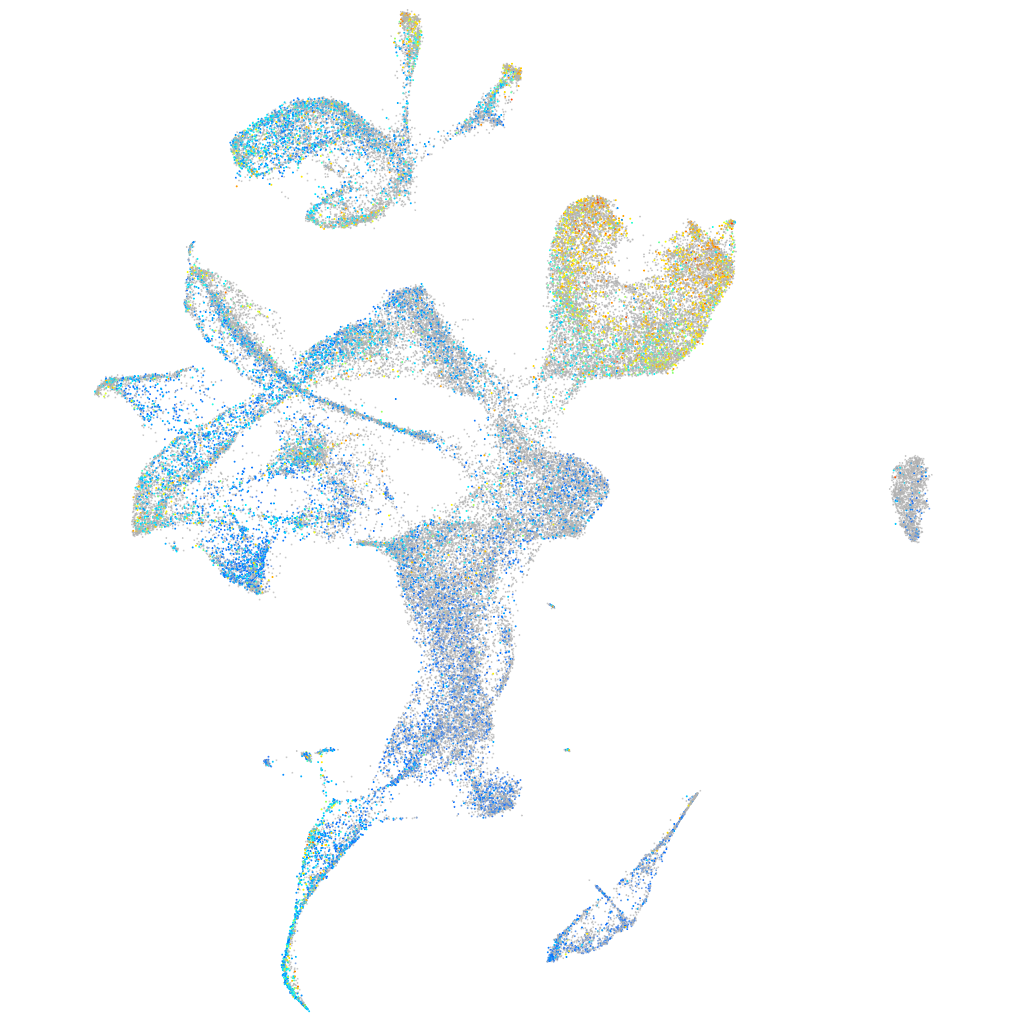WBP2 N-terminal like
ZFIN 
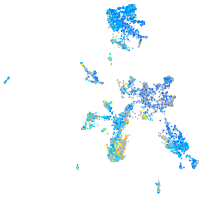
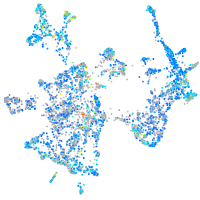
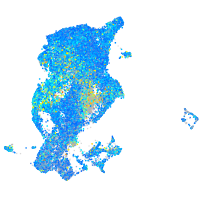
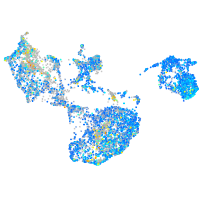
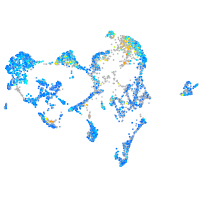
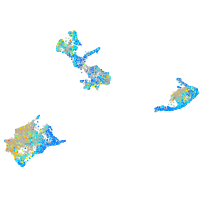
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1e1b | 0.200 | si:ch211-222l21.1 | -0.179 |
| atp6v1g1 | 0.181 | hmga1a | -0.177 |
| atp6v0cb | 0.170 | hmgb2b | -0.171 |
| gapdhs | 0.161 | rplp1 | -0.148 |
| snap25b | 0.160 | hmgn2 | -0.147 |
| ckbb | 0.156 | rplp2l | -0.147 |
| atp6ap2 | 0.155 | pcna | -0.146 |
| id2a | 0.154 | stmn1a | -0.145 |
| rnasekb | 0.152 | rrm1 | -0.144 |
| calm1a | 0.152 | mki67 | -0.144 |
| tpi1b | 0.149 | rps12 | -0.142 |
| si:ch211-195b13.1 | 0.147 | seta | -0.138 |
| ppdpfb | 0.146 | ran | -0.137 |
| atp6v1h | 0.145 | anp32b | -0.135 |
| rgra | 0.144 | si:dkey-151g10.6 | -0.135 |
| tyrp1b | 0.144 | lbr | -0.135 |
| atpv0e2 | 0.142 | ccna2 | -0.134 |
| dct | 0.142 | snrpb | -0.134 |
| pmela | 0.142 | nasp | -0.133 |
| pmelb | 0.142 | chaf1a | -0.131 |
| calm1b | 0.141 | dek | -0.131 |
| gpr143 | 0.140 | ccnd1 | -0.130 |
| tob1b | 0.139 | rpl9 | -0.130 |
| rlbp1b | 0.138 | hmgb2a | -0.130 |
| tspan10 | 0.138 | snrpd1 | -0.129 |
| rgrb | 0.137 | rps28 | -0.128 |
| sypb | 0.137 | rpl35a | -0.128 |
| sparc | 0.135 | mcm7 | -0.127 |
| oca2 | 0.134 | banf1 | -0.127 |
| msnb | 0.134 | rpsa | -0.126 |
| fabp11b | 0.133 | h2afvb | -0.126 |
| slc45a2 | 0.133 | cx43.4 | -0.125 |
| syngr1a | 0.133 | lig1 | -0.124 |
| tyrp1a | 0.133 | rpl23a | -0.124 |
| bace2 | 0.132 | rpl12 | -0.123 |