WASH complex subunit 3
ZFIN 
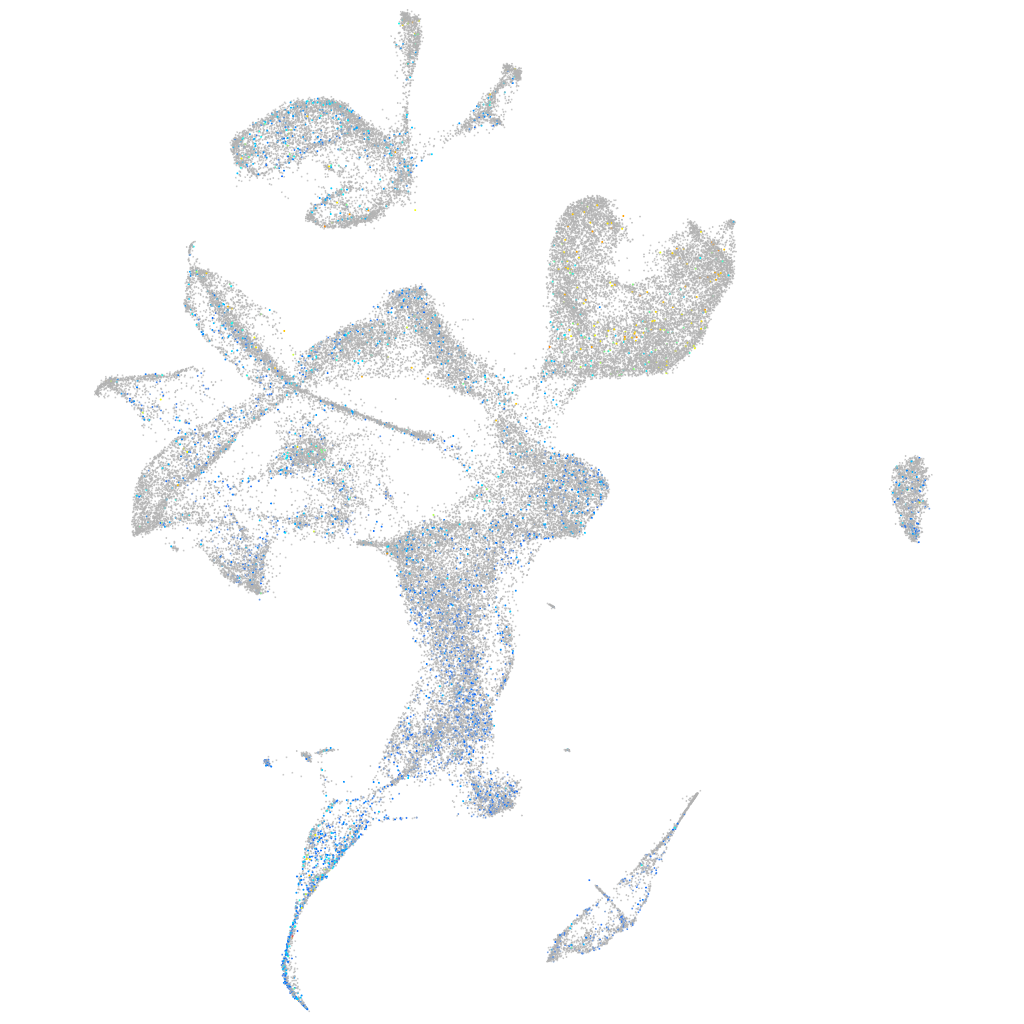
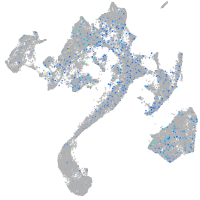
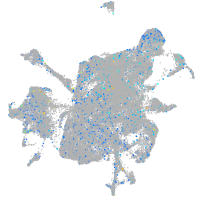
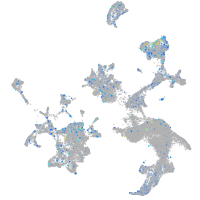
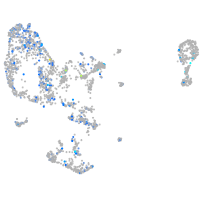
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpr143 | 0.154 | ptmab | -0.072 |
| dct | 0.153 | si:ch73-1a9.3 | -0.054 |
| tspan36 | 0.152 | hmgn6 | -0.049 |
| slc45a2 | 0.148 | syt5b | -0.046 |
| tyrp1b | 0.146 | ptmaa | -0.043 |
| pmela | 0.146 | sypb | -0.041 |
| pmelb | 0.142 | neurod4 | -0.039 |
| fabp11b | 0.136 | crx | -0.038 |
| tyrp1a | 0.136 | vsx1 | -0.038 |
| cracr2ab | 0.135 | nrn1lb | -0.035 |
| rab38 | 0.134 | epb41a | -0.035 |
| tspan10 | 0.133 | snap25b | -0.034 |
| bace2 | 0.132 | rs1a | -0.034 |
| qdpra | 0.131 | cplx4a | -0.033 |
| oca2 | 0.129 | samsn1a | -0.033 |
| agtrap | 0.128 | ckbb | -0.033 |
| slc24a5 | 0.127 | anp32e | -0.032 |
| triobpa | 0.125 | calb2b | -0.031 |
| pttg1ipb | 0.125 | fabp7a | -0.030 |
| rgrb | 0.124 | pvalb1 | -0.030 |
| rab32a | 0.124 | vamp1 | -0.030 |
| mitfa | 0.123 | actc1b | -0.030 |
| zgc:110591 | 0.123 | pvalb2 | -0.029 |
| gstp1 | 0.122 | npdc1a | -0.029 |
| FP085398.1 | 0.120 | ppp1r1b | -0.029 |
| tm6sf2 | 0.119 | gapdhs | -0.028 |
| msnb | 0.118 | mpp6b | -0.028 |
| rnaseka | 0.117 | tuba1c | -0.028 |
| fgfbp1a | 0.115 | myhz1.1 | -0.028 |
| tmem98 | 0.115 | cadm3 | -0.028 |
| tyr | 0.115 | neurod1 | -0.028 |
| col4a5 | 0.114 | stxbp1b | -0.028 |
| trpm1b | 0.114 | gnb3a | -0.028 |
| pnp4a | 0.113 | stx3a | -0.027 |
| rlbp1b | 0.113 | gpm6aa | -0.027 |