WASP family member 2
ZFIN 
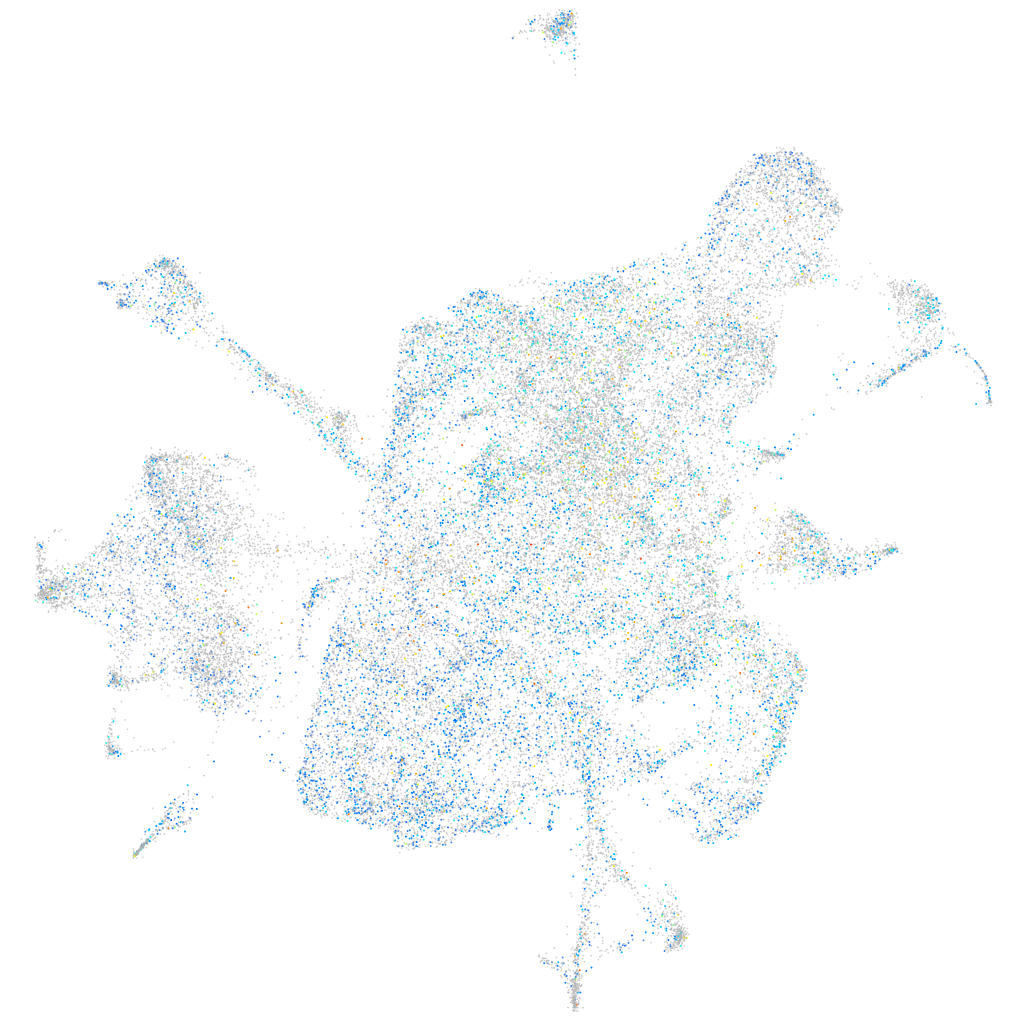
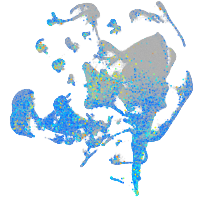
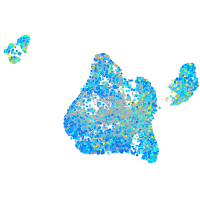
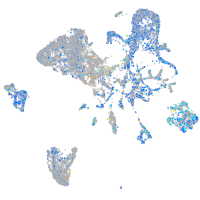
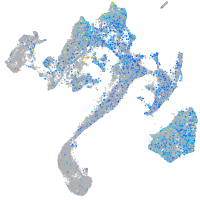
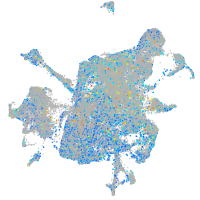
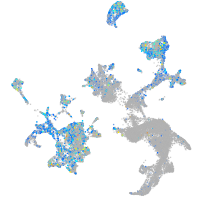
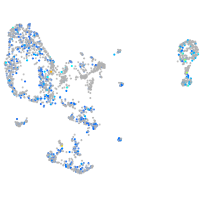
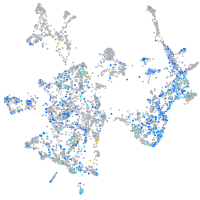
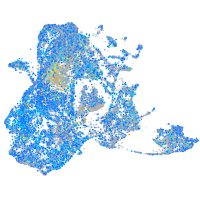
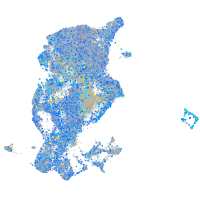
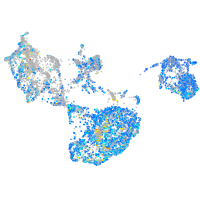
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tpm4a | 0.100 | gpm6aa | -0.056 |
| actb1 | 0.086 | ckbb | -0.047 |
| zgc:153867 | 0.083 | elavl3 | -0.046 |
| gng5 | 0.080 | mdkb | -0.045 |
| myl12.1 | 0.079 | stmn1b | -0.044 |
| krt18b | 0.077 | gng3 | -0.044 |
| pmp22a | 0.076 | otos | -0.042 |
| msna | 0.076 | pvalb2 | -0.040 |
| actb2 | 0.075 | rtn1a | -0.040 |
| mfap2 | 0.074 | apoa2 | -0.040 |
| cnn2 | 0.074 | cspg5a | -0.039 |
| plscr3b | 0.073 | tuba1c | -0.039 |
| sept2 | 0.073 | col11a2 | -0.039 |
| cfl1 | 0.072 | pvalb1 | -0.038 |
| sri | 0.069 | rtn1b | -0.037 |
| akap12b | 0.068 | atp6v0cb | -0.037 |
| sept10 | 0.068 | apoa1b | -0.036 |
| rac1a | 0.067 | fez1 | -0.036 |
| si:ch211-286o17.1 | 0.065 | matn1 | -0.036 |
| krt8 | 0.065 | snap25b | -0.036 |
| ahnak | 0.065 | col2a1a | -0.035 |
| ywhaz | 0.063 | epyc | -0.035 |
| marcksl1a | 0.062 | gnao1a | -0.035 |
| krt18a.1 | 0.062 | myt1b | -0.035 |
| flot2a | 0.062 | tmsb | -0.034 |
| flot1b | 0.062 | fgfbp2b | -0.034 |
| ubc | 0.062 | stx1b | -0.034 |
| klf6a | 0.062 | gpm6ab | -0.034 |
| si:dkey-261h17.1 | 0.062 | CU467822.1 | -0.033 |
| colec12 | 0.061 | sncb | -0.033 |
| tuba8l3 | 0.061 | CELA1 (1 of many) | -0.032 |
| rap1b | 0.060 | col9a1a | -0.032 |
| cdc42l | 0.058 | si:dkey-56f14.7 | -0.032 |
| emp2 | 0.058 | cthrc1b | -0.032 |
| pdia3 | 0.058 | tnxba | -0.032 |