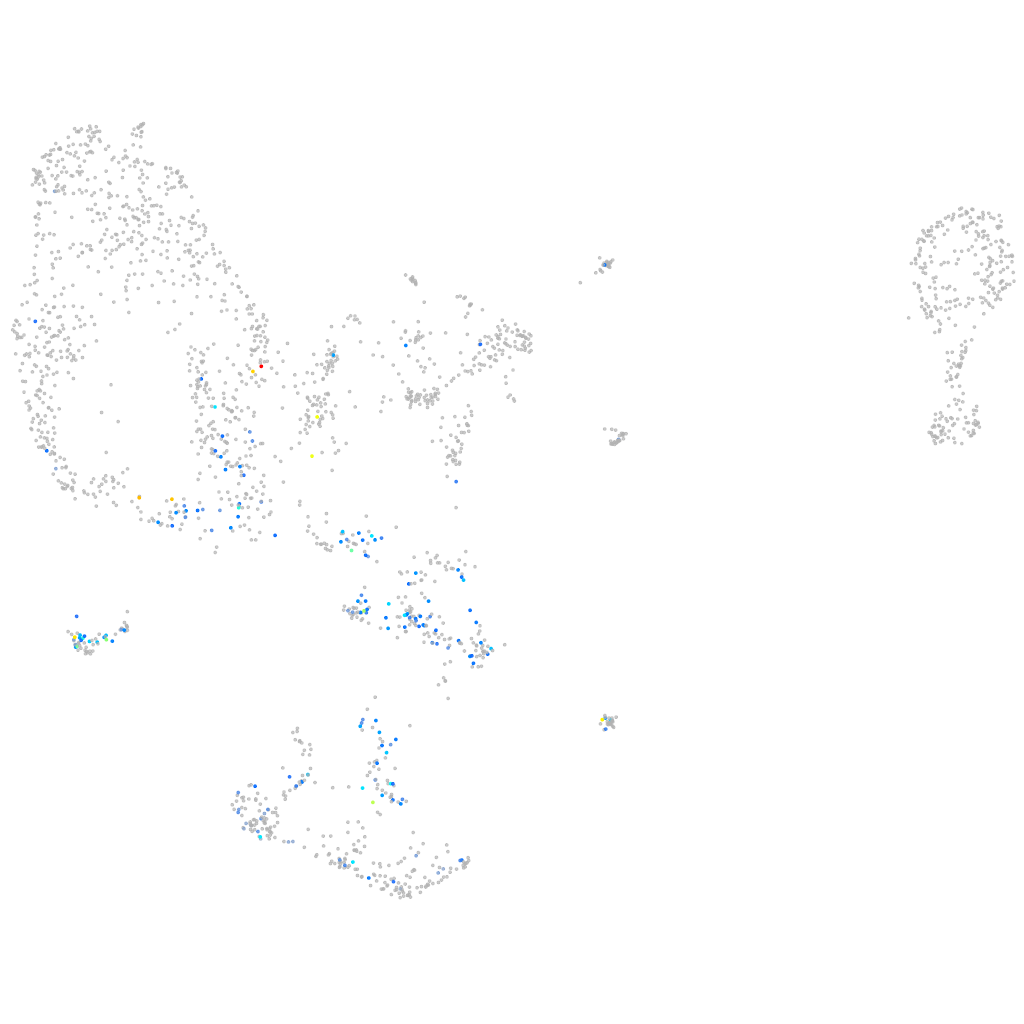
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 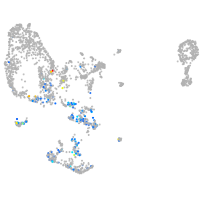 |
neural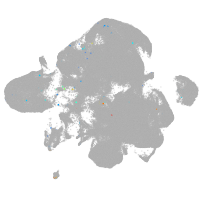 |
spinal cord / glia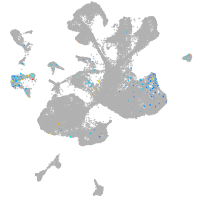 |
eye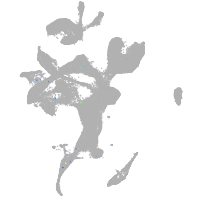 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| spaca9 | 0.326 | gcshb | -0.148 |
| tex9 | 0.313 | gapdh | -0.144 |
| rsph9 | 0.299 | si:dkey-33i11.9 | -0.144 |
| ccdc151 | 0.296 | phyhd1 | -0.143 |
| dnali1 | 0.288 | agxtb | -0.139 |
| mycbp | 0.287 | aldob | -0.138 |
| si:ch211-248e11.2 | 0.285 | gstt1a | -0.137 |
| casc1 | 0.285 | gpt2l | -0.136 |
| pih1d3 | 0.281 | pnp6 | -0.133 |
| ccdc114 | 0.276 | grhprb | -0.132 |
| cfap45 | 0.275 | upb1 | -0.131 |
| ift22 | 0.274 | hoga1 | -0.127 |
| spata18 | 0.273 | rps6 | -0.126 |
| cfap20 | 0.273 | hao1 | -0.126 |
| spag6 | 0.273 | haao | -0.124 |
| daw1 | 0.271 | rps18 | -0.123 |
| zgc:112416 | 0.270 | cx32.3 | -0.123 |
| meig1 | 0.270 | g6pca.2 | -0.122 |
| catip | 0.268 | dpys | -0.122 |
| tekt1 | 0.267 | dhrs9 | -0.121 |
| ccdc173 | 0.266 | fbp1b | -0.120 |
| efcab1 | 0.266 | rgn | -0.120 |
| capsla | 0.265 | si:ch211-139a5.9 | -0.119 |
| tekt2 | 0.264 | slc22a4 | -0.118 |
| si:ch211-93n23.7 | 0.264 | si:ch211-201h21.5 | -0.118 |
| efcab2 | 0.262 | slc5a12 | -0.118 |
| cfap52 | 0.261 | slc26a6l | -0.117 |
| cdc25d | 0.260 | nit2 | -0.117 |
| spata4 | 0.260 | gamt | -0.116 |
| si:dkey-148f10.4 | 0.260 | cat | -0.116 |
| si:ch211-163l21.7 | 0.259 | fahd1 | -0.116 |
| ift27 | 0.259 | aldh7a1 | -0.116 |
| erich2 | 0.258 | l2hgdh | -0.115 |
| crocc2 | 0.255 | slc47a4 | -0.115 |
| LOC103911624 | 0.254 | ctsla | -0.115 |