V-set and transmembrane domain containing 2 like
ZFIN 
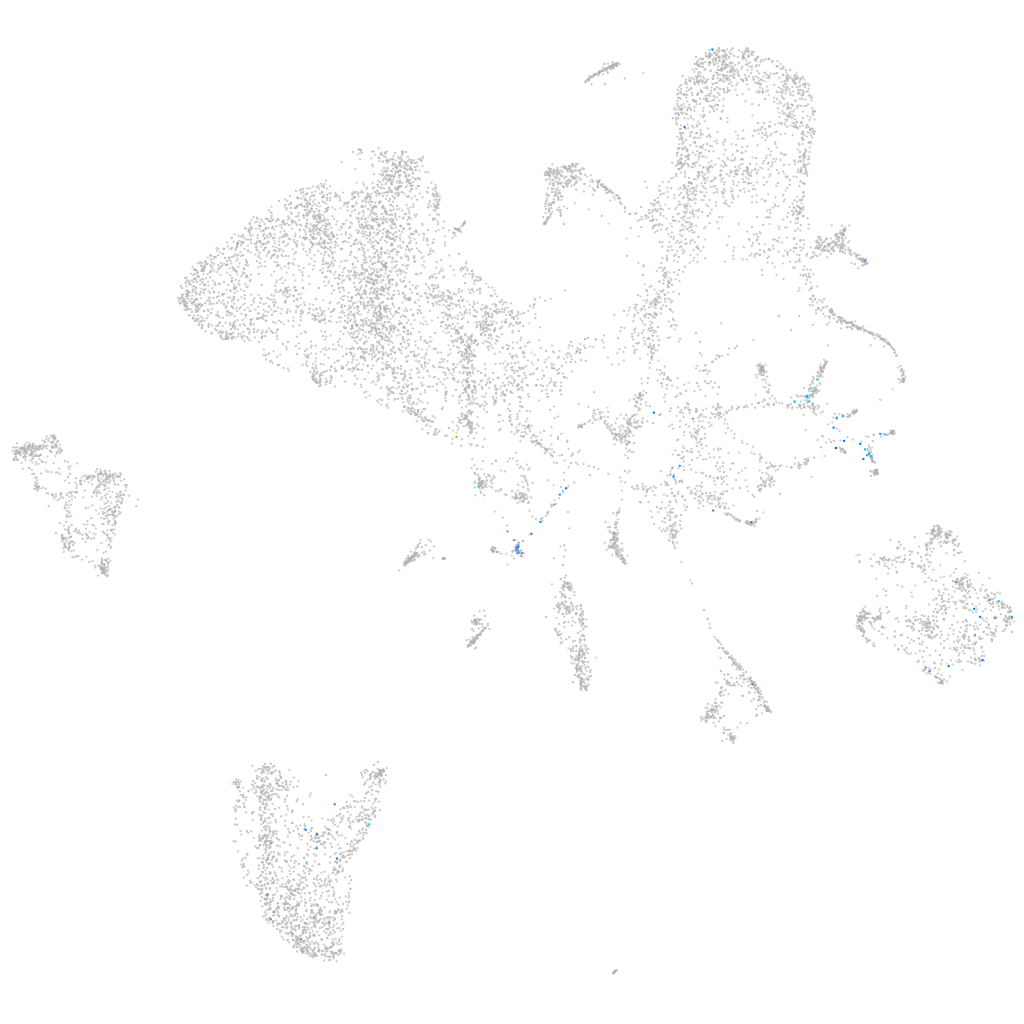
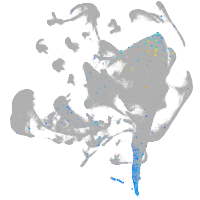
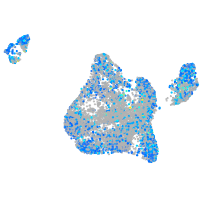
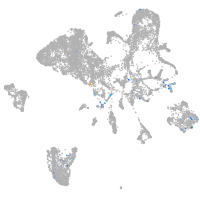
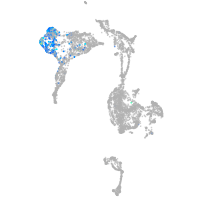
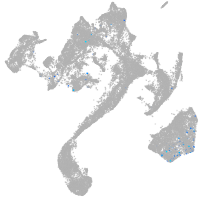
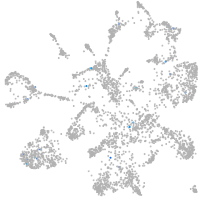
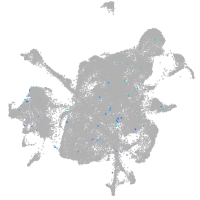
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gabrz | 0.309 | gstt1a | -0.075 |
| XLOC-025875 | 0.267 | fbp1b | -0.070 |
| XLOC-002533 | 0.234 | gapdh | -0.070 |
| si:ch73-290k24.6 | 0.214 | ahcy | -0.069 |
| nlrp16 | 0.206 | apoa4b.1 | -0.068 |
| CABZ01087502.1 | 0.205 | gamt | -0.068 |
| gfra2b | 0.201 | aldob | -0.068 |
| XLOC-036961 | 0.197 | apoc2 | -0.067 |
| gabrd | 0.195 | apoa1b | -0.066 |
| XLOC-005257 | 0.193 | afp4 | -0.066 |
| rprmb | 0.193 | sdr16c5b | -0.063 |
| apba2a | 0.186 | gstp1 | -0.063 |
| kcnn1a | 0.185 | scp2a | -0.062 |
| sst1.1 | 0.179 | gcshb | -0.062 |
| adgrb3 | 0.178 | glud1b | -0.061 |
| slc35g2b | 0.176 | rdh1 | -0.061 |
| kcnj11 | 0.176 | sult2st2 | -0.060 |
| zgc:152863 | 0.175 | eno3 | -0.059 |
| admb | 0.174 | gpx4a | -0.059 |
| dock3 | 0.172 | gsta.1 | -0.058 |
| LOC103908906 | 0.172 | tdo2a | -0.058 |
| phf1 | 0.168 | shmt1 | -0.058 |
| kcnk9 | 0.166 | mgst1.2 | -0.058 |
| si:ch211-288d18.1 | 0.165 | aldh7a1 | -0.057 |
| BX890591.1 | 0.164 | si:dkey-16p21.8 | -0.057 |
| tspan7b | 0.164 | haao | -0.056 |
| LOC103909072 | 0.163 | suclg2 | -0.056 |
| pcdh1g32 | 0.162 | srd5a2a | -0.056 |
| pcdh2ab11 | 0.162 | fabp2 | -0.056 |
| cacna2d1a | 0.161 | ugt1a7 | -0.056 |
| CABZ01043955.1 | 0.161 | serpina1l | -0.056 |
| elmod1 | 0.160 | apobb.1 | -0.055 |
| lrrtm2 | 0.159 | serpina1 | -0.055 |
| XLOC-011085 | 0.159 | akr7a3 | -0.055 |
| rem2 | 0.158 | rmdn1 | -0.055 |