VPS18 core subunit of CORVET and HOPS complexes
ZFIN 
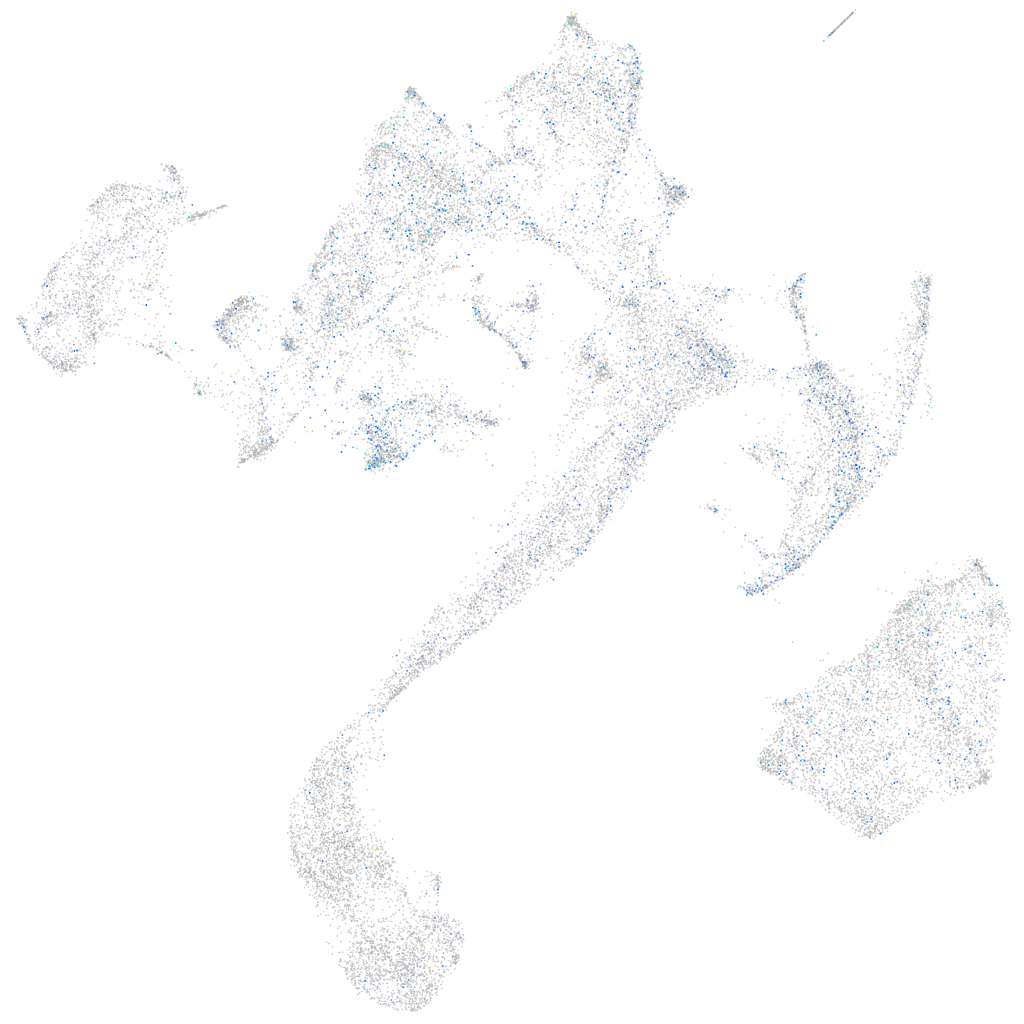
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR381646.1 | 0.081 | acta1b | -0.057 |
| cfl1 | 0.076 | tnnc2 | -0.056 |
| ptmaa | 0.076 | aldoab | -0.053 |
| h3f3a | 0.075 | tpma | -0.052 |
| actb2 | 0.072 | actn3b | -0.052 |
| sncb | 0.071 | tmem38a | -0.051 |
| ppiab | 0.071 | srl | -0.051 |
| stxbp1a | 0.071 | neb | -0.050 |
| ywhabl | 0.070 | actn3a | -0.050 |
| h3f3c | 0.069 | gapdh | -0.050 |
| stx1b | 0.067 | smyd1a | -0.050 |
| tubb4b | 0.067 | atp2a1 | -0.050 |
| actb1 | 0.066 | ckmb | -0.049 |
| zgc:153867 | 0.065 | ldb3a | -0.049 |
| hnrnpa0l | 0.064 | ldb3b | -0.049 |
| stmn1b | 0.064 | tmod4 | -0.049 |
| calr | 0.064 | actc1b | -0.049 |
| tuba1c | 0.064 | hhatla | -0.049 |
| zgc:65894 | 0.064 | ak1 | -0.049 |
| CR974456.1 | 0.063 | ckma | -0.048 |
| elavl4 | 0.063 | ttn.1 | -0.048 |
| atp6v1e1b | 0.063 | mybphb | -0.048 |
| vamp2 | 0.063 | pgam2 | -0.047 |
| ap2m1a | 0.063 | ttn.2 | -0.047 |
| gnao1a | 0.063 | acta1a | -0.047 |
| tmsb4x | 0.063 | tnnt3a | -0.047 |
| gng3 | 0.062 | nme2b.2 | -0.047 |
| hmgn3 | 0.062 | XLOC-006515 | -0.046 |
| ubc | 0.062 | casq2 | -0.046 |
| epb41a | 0.062 | myl1 | -0.046 |
| gapdhs | 0.062 | XLOC-025819 | -0.046 |
| marcksl1a | 0.062 | si:ch211-255p10.3 | -0.046 |
| eno2 | 0.061 | myom1a | -0.045 |
| ppib | 0.061 | si:ch73-367p23.2 | -0.045 |
| atp6ap1a | 0.061 | CABZ01078594.1 | -0.045 |