vascular endothelial growth factor Bb
ZFIN 
Other cell groups


all cells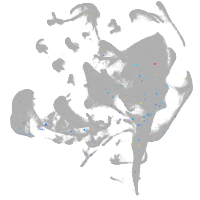 |
blastomeres |
PGCs |
endoderm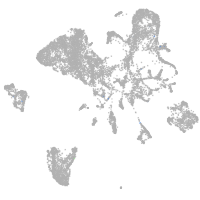 |
axial mesoderm |
muscle 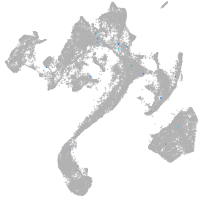 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 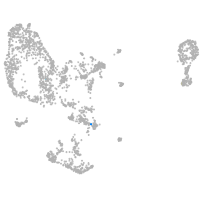 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line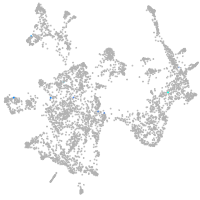 |
epidermis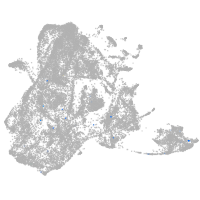 |
periderm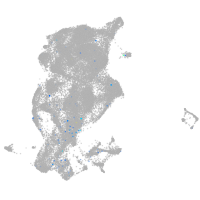 |
fin |
ionocytes / mucous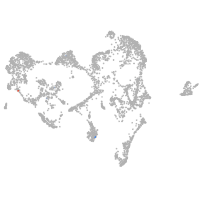 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tgm1l4 | 0.033 | ckbb | -0.013 |
| capn9 | 0.030 | rtn1a | -0.012 |
| si:ch73-347e22.8 | 0.028 | gpm6ab | -0.011 |
| zgc:153665 | 0.028 | pvalb1 | -0.011 |
| si:cabz01007794.1 | 0.027 | pvalb2 | -0.011 |
| zgc:174938 | 0.027 | stmn1b | -0.011 |
| ANXA1 (1 of many) | 0.027 | vdac3 | -0.011 |
| agr1 | 0.026 | atp6v0cb | -0.010 |
| mgst2 | 0.026 | elavl3 | -0.010 |
| pcna | 0.026 | gapdhs | -0.010 |
| rab25a | 0.026 | gng3 | -0.010 |
| si:ch211-157c3.4 | 0.026 | gpm6aa | -0.010 |
| anxa1b | 0.025 | jpt1b | -0.010 |
| anxa1c | 0.025 | si:dkeyp-75h12.5 | -0.010 |
| cldn23a | 0.025 | actc1b | -0.009 |
| evplb | 0.025 | atp6v1e1b | -0.009 |
| lye | 0.025 | col1a2 | -0.009 |
| mcm4 | 0.025 | dpysl3 | -0.009 |
| si:ch73-78o10.1 | 0.025 | elavl4 | -0.009 |
| si:dkey-87o1.2 | 0.025 | fez1 | -0.009 |
| ftr83 | 0.025 | gap43 | -0.009 |
| rrm2 | 0.025 | gng2 | -0.009 |
| acer1 | 0.024 | mylpfa | -0.009 |
| ftr86 | 0.024 | rnasekb | -0.009 |
| hsd17b12a | 0.024 | rtn1b | -0.009 |
| krt92 | 0.024 | scrt2 | -0.009 |
| LOC100535067 | 0.024 | sncb | -0.009 |
| ppl | 0.024 | sparc | -0.009 |
| rrm1 | 0.024 | stmn2a | -0.009 |
| si:ch73-204p21.2 | 0.024 | stx1b | -0.009 |
| stx11b.1 | 0.024 | tmsb | -0.009 |
| XLOC-027129 | 0.024 | ywhag2 | -0.009 |
| zgc:194839 | 0.024 | zc4h2 | -0.009 |
| cldne | 0.023 | col1a1a | -0.008 |
| cst14b.1 | 0.023 | col1a1b | -0.008 |




