voltage-dependent anion channel 1
ZFIN 
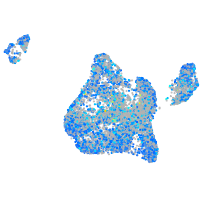
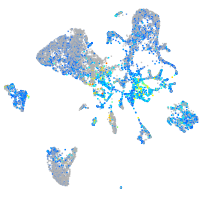
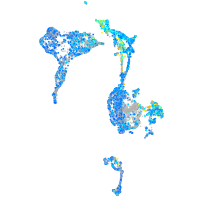
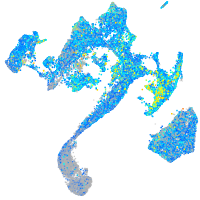
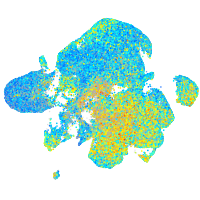
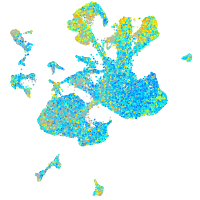
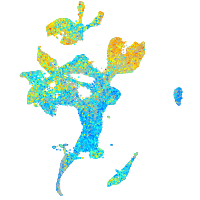
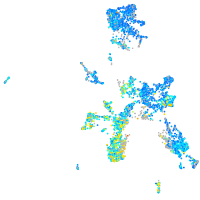
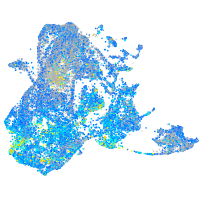
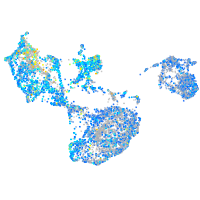
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.367 | rbp4 | -0.188 |
| tpi1b | 0.345 | NC-002333.4 | -0.168 |
| gpia | 0.345 | podxl | -0.153 |
| eno3 | 0.324 | CR383676.1 | -0.143 |
| pkma | 0.317 | rbpms2b | -0.141 |
| pgk1 | 0.316 | krt94 | -0.136 |
| fbp2 | 0.315 | aqp1a.1 | -0.130 |
| pgam1a | 0.315 | CABZ01072614.1 | -0.129 |
| ckba | 0.314 | nid1a | -0.129 |
| csrp1b | 0.313 | ednraa | -0.128 |
| ldha | 0.309 | rhag | -0.128 |
| aldocb | 0.306 | jam2b | -0.125 |
| pfkpa | 0.298 | fabp11a | -0.125 |
| ybx1 | 0.293 | LOC110439372 | -0.125 |
| slc16a3 | 0.285 | colec12 | -0.123 |
| acta2 | 0.281 | notch3 | -0.122 |
| tpm2 | 0.279 | akap12b | -0.120 |
| eno1a | 0.278 | ephb3 | -0.117 |
| pfn1 | 0.278 | mfap5 | -0.116 |
| atp5f1b | 0.277 | COX3 | -0.113 |
| ckbb | 0.275 | bnc2 | -0.112 |
| vdac2 | 0.273 | zgc:158463 | -0.112 |
| myl6 | 0.271 | fgfr2 | -0.112 |
| cald1b | 0.270 | col18a1a | -0.110 |
| atp5mc1 | 0.267 | c1qtnf6a | -0.107 |
| myl9a | 0.265 | NC-002333.17 | -0.106 |
| acta1a | 0.259 | c6 | -0.105 |
| tuba8l2 | 0.258 | col12a1a | -0.105 |
| tpm1 | 0.255 | nrp1a | -0.103 |
| slc2a1b | 0.254 | wt1a | -0.102 |
| cox6b1 | 0.253 | nid1b | -0.102 |
| zgc:153867 | 0.253 | f10 | -0.102 |
| tagln | 0.252 | zeb2a | -0.101 |
| calm3b | 0.250 | pdgfrb | -0.100 |
| bricd5 | 0.249 | gata5 | -0.098 |