vascular cell adhesion molecule 1b
ZFIN 
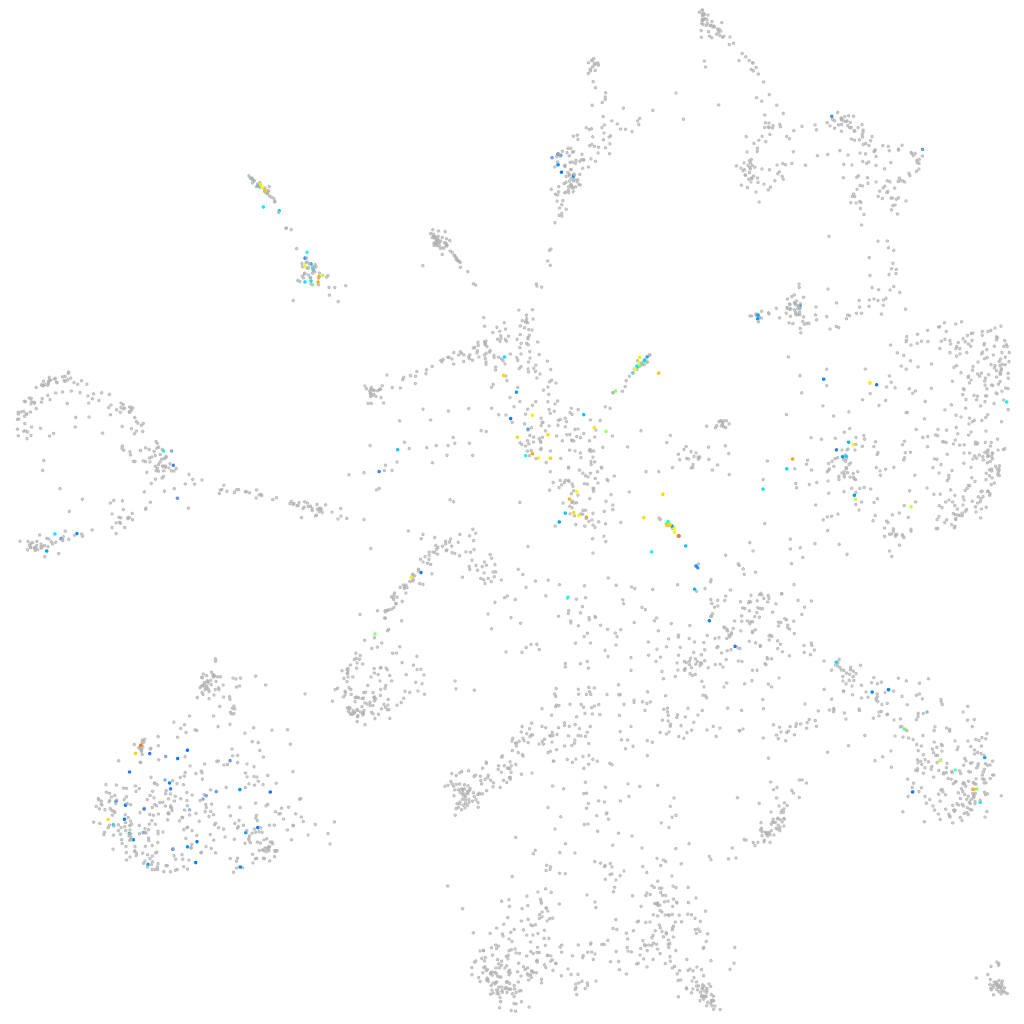
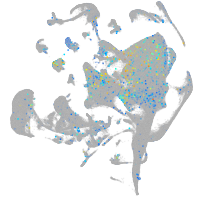
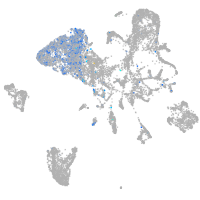
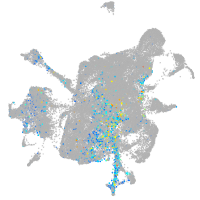
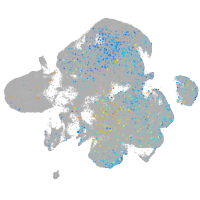
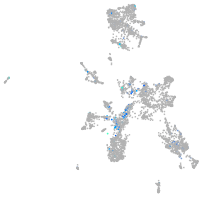
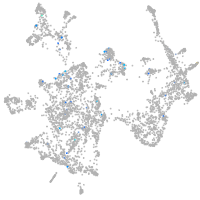
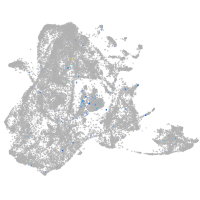
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rgs5b | 0.271 | myl9a | -0.130 |
| ndufa4l2a | 0.241 | foxp4 | -0.113 |
| pdgfrb | 0.236 | hand2 | -0.109 |
| tlx1 | 0.229 | ckba | -0.108 |
| pcdh18a | 0.222 | acta2 | -0.106 |
| atp1b4 | 0.210 | foxf1 | -0.102 |
| il15 | 0.200 | hmga2 | -0.100 |
| tril | 0.190 | col6a2 | -0.097 |
| XLOC-016094 | 0.185 | cavin1b | -0.093 |
| si:ch211-195b13.1 | 0.184 | mylkb | -0.091 |
| adra2c | 0.183 | ntn5 | -0.091 |
| atp1a1b | 0.183 | myl6 | -0.088 |
| FO704741.1 | 0.182 | tpm2 | -0.086 |
| zeb2a | 0.181 | csrp1a | -0.086 |
| cx39.4 | 0.179 | col6a1 | -0.086 |
| rasl12 | 0.178 | si:dkeyp-66d1.7 | -0.085 |
| spns2 | 0.176 | ociad2 | -0.084 |
| notch3 | 0.176 | col4a5 | -0.082 |
| ramp2 | 0.174 | csrp1b | -0.082 |
| sh3d21 | 0.170 | col1a1b | -0.081 |
| nkx2.7 | 0.170 | col4a6 | -0.081 |
| epas1a | 0.169 | lmod1b | -0.080 |
| slc29a1a | 0.167 | si:ch211-62a1.3 | -0.079 |
| cox4i2 | 0.163 | flna | -0.077 |
| pag1 | 0.163 | desmb | -0.077 |
| bgnb | 0.160 | zgc:114188 | -0.075 |
| ntrk2a | 0.158 | cavin2b | -0.074 |
| FO704758.1 | 0.157 | elovl1b | -0.074 |
| rasl11a | 0.156 | cald1b | -0.073 |
| wfikkn2a | 0.156 | cnn1b | -0.073 |
| prox2 | 0.156 | foxf2a | -0.073 |
| tm4sf18 | 0.155 | gucy1a1 | -0.072 |
| kcnj8 | 0.155 | rpl24 | -0.072 |
| agtr2 | 0.154 | tes | -0.071 |
| ebf1a | 0.154 | hlx1 | -0.070 |