VANGL planar cell polarity protein 2
ZFIN 
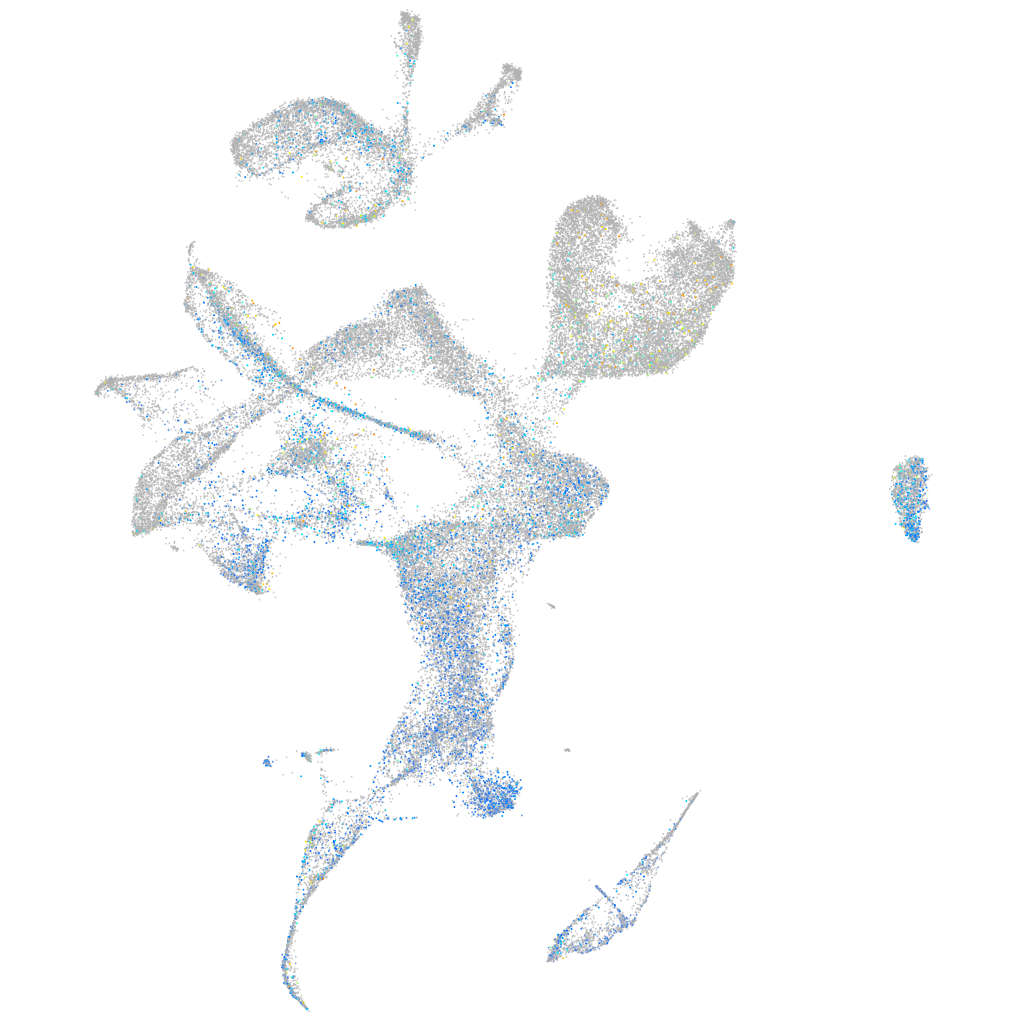
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ctnnb1 | 0.102 | snap25b | -0.096 |
| id1 | 0.101 | gapdhs | -0.091 |
| si:ch211-222l21.1 | 0.101 | syt5b | -0.091 |
| eef1a1l1 | 0.097 | atp6v0cb | -0.084 |
| eef2b | 0.094 | ckbb | -0.081 |
| nop58 | 0.094 | sypb | -0.081 |
| hspb1 | 0.093 | atpv0e2 | -0.077 |
| bzw1b | 0.093 | rnasekb | -0.072 |
| rpl9 | 0.091 | crx | -0.072 |
| ctsla | 0.091 | sh3gl2a | -0.071 |
| si:dkey-56m19.5 | 0.089 | rs1a | -0.070 |
| tspan7 | 0.089 | atp2b1b | -0.067 |
| dkc1 | 0.089 | cplx4a | -0.066 |
| eif5a2 | 0.089 | neurod1 | -0.064 |
| npm1a | 0.088 | otx5 | -0.064 |
| fbl | 0.088 | cnrip1b | -0.062 |
| sfrp1a | 0.088 | atp6v1e1b | -0.062 |
| alcamb | 0.088 | lin7a | -0.061 |
| tuba8l4 | 0.087 | nptnb | -0.061 |
| khdrbs1a | 0.087 | calb2b | -0.060 |
| vamp3 | 0.086 | anp32e | -0.060 |
| nop56 | 0.085 | pvalb1 | -0.059 |
| nop2 | 0.085 | atp6v1g1 | -0.058 |
| cx43.4 | 0.085 | aldocb | -0.057 |
| her6 | 0.085 | eno1a | -0.057 |
| ybx1 | 0.085 | pvalb2 | -0.057 |
| nhp2 | 0.085 | tulp1a | -0.056 |
| marcksb | 0.085 | tpi1b | -0.056 |
| syncrip | 0.084 | vamp2 | -0.056 |
| ranbp1 | 0.084 | pacsin1a | -0.056 |
| apoeb | 0.083 | gnb5b | -0.055 |
| ppiaa | 0.083 | stxbp1b | -0.055 |
| pprc1 | 0.083 | si:dkey-16p21.8 | -0.055 |
| akap12b | 0.083 | gngt2a | -0.054 |
| gng5 | 0.083 | ckmt2a | -0.054 |