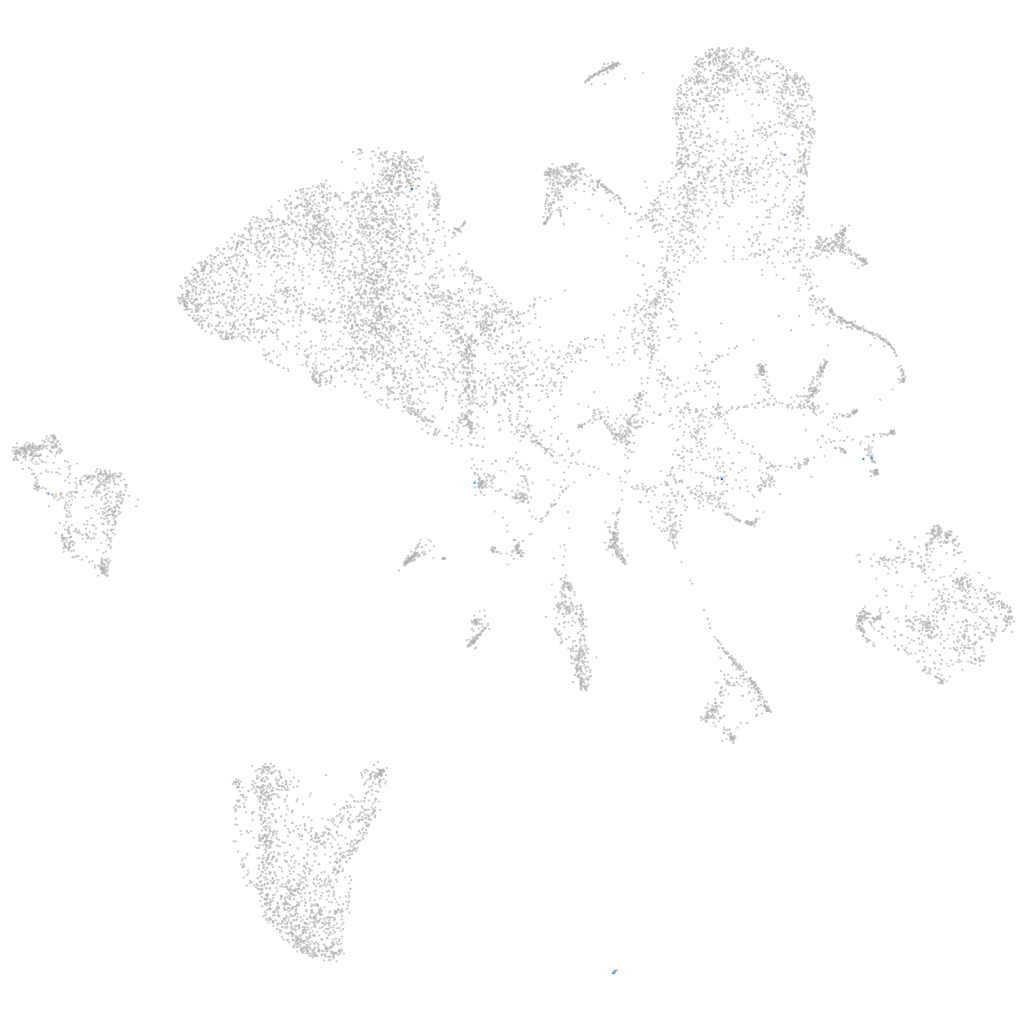
ubiquitin specific peptidase 54b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| BX511024.1 | 0.558 | aldob | -0.029 |
| LOC108192083 | 0.558 | rpl6 | -0.029 |
| CU657925.1 | 0.549 | gapdh | -0.025 |
| nts | 0.517 | rps4x | -0.022 |
| LOC108190036 | 0.507 | dap | -0.021 |
| tmem229a | 0.460 | hspe1 | -0.021 |
| RNF157 | 0.396 | prelid1a | -0.020 |
| LOC559637 | 0.390 | rpsa | -0.020 |
| atp8b2 | 0.360 | aldh9a1a.1 | -0.020 |
| fbxo41 | 0.349 | aldh7a1 | -0.019 |
| pcdh1gc6 | 0.340 | zgc:162730 | -0.019 |
| slc24a4b | 0.333 | lgals2b | -0.019 |
| nptxra | 0.324 | gamt | -0.019 |
| neto1l | 0.313 | aqp12 | -0.019 |
| CABZ01092282.1 | 0.303 | slc16a1b | -0.018 |
| CABZ01064248.1 | 0.263 | cldn15lb | -0.018 |
| si:dkey-238f9.1 | 0.260 | chchd10 | -0.018 |
| glra3 | 0.257 | fdx1 | -0.018 |
| grm8a | 0.256 | tmem214 | -0.018 |
| irx2a | 0.256 | gstz1 | -0.018 |
| KIRREL3 | 0.255 | gchfr | -0.018 |
| NRXN2 | 0.255 | fbp1b | -0.018 |
| si:dkey-27p18.5 | 0.251 | eno3 | -0.018 |
| lbx1a | 0.245 | sb:cb1058 | -0.018 |
| CABZ01050944.1 | 0.241 | ppdpfa | -0.018 |
| cadm1a | 0.235 | gstr | -0.018 |
| trpc1 | 0.235 | pgk1 | -0.017 |
| XLOC-041467 | 0.227 | gstt1a | -0.017 |
| camk1a | 0.226 | gnmt | -0.017 |
| CABZ01073166.1 | 0.225 | pgm1 | -0.017 |
| CABZ01053748.1 | 0.221 | slc16a10 | -0.017 |
| trarg1a | 0.221 | mat1a | -0.017 |
| si:dkey-56f14.4 | 0.217 | gpx4a | -0.017 |
| dmrt3a | 0.214 | acaa2 | -0.017 |
| FP017161.1 | 0.214 | pklr | -0.017 |