ubiquinol-cytochrome c reductase core protein 2a
ZFIN 
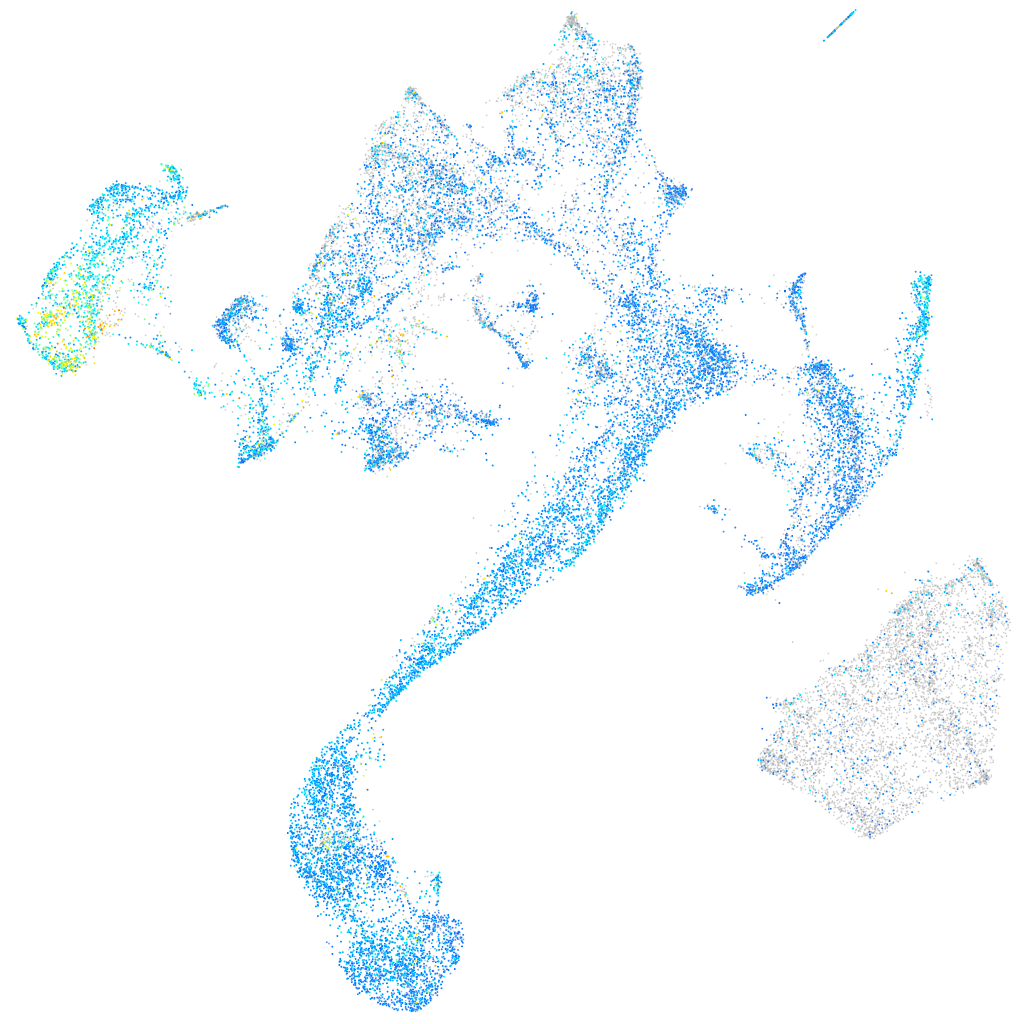
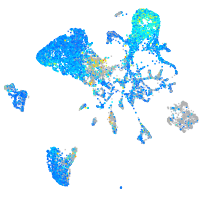
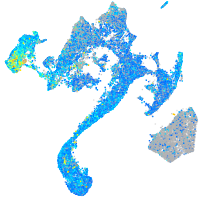
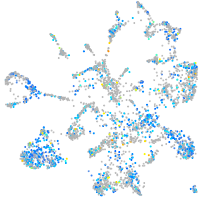
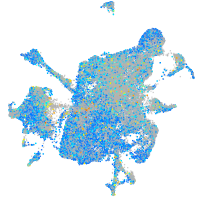
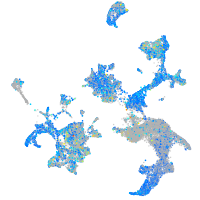
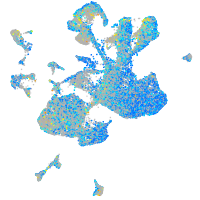
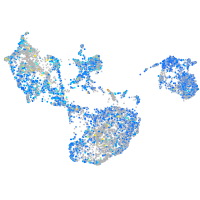
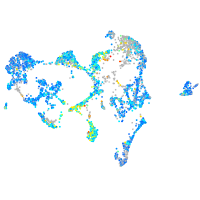
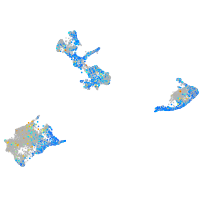
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5f1b | 0.521 | ptmab | -0.477 |
| suclg1 | 0.505 | si:ch73-1a9.3 | -0.466 |
| aldoab | 0.499 | si:ch211-222l21.1 | -0.427 |
| atp5if1b | 0.497 | pabpc1a | -0.424 |
| atp5meb | 0.490 | anp32a | -0.402 |
| atp5mc3b | 0.487 | hmgb2a | -0.397 |
| ttn.2 | 0.486 | hmgb2b | -0.386 |
| ttn.1 | 0.484 | acin1a | -0.385 |
| cox7a2a | 0.484 | hsp90ab1 | -0.383 |
| srl | 0.482 | hnrnpa1b | -0.381 |
| tmem38a | 0.478 | si:ch73-281n10.2 | -0.381 |
| vdac2 | 0.477 | cirbpa | -0.380 |
| atp5mc1 | 0.472 | nucks1a | -0.378 |
| actc1b | 0.471 | seta | -0.377 |
| atp5fa1 | 0.471 | marcksb | -0.368 |
| desma | 0.467 | cx43.4 | -0.367 |
| mdh2 | 0.466 | anp32b | -0.363 |
| acta1a | 0.463 | hnrnpub | -0.357 |
| cox6a1 | 0.456 | hmga1a | -0.356 |
| atp2a1 | 0.454 | hnrnpa0b | -0.356 |
| ak1 | 0.453 | hnrnpaba | -0.356 |
| cav3 | 0.448 | hnrnpa1a | -0.355 |
| casq2 | 0.448 | si:ch211-288g17.3 | -0.355 |
| COX5B | 0.445 | marcksl1b | -0.354 |
| eno3 | 0.445 | h3f3d | -0.350 |
| cox5aa | 0.444 | hnrnpm | -0.349 |
| mt-nd1 | 0.443 | tuba8l4 | -0.348 |
| atp5pd | 0.443 | cdx4 | -0.348 |
| gapdh | 0.443 | khdrbs1a | -0.346 |
| ckmb | 0.440 | hmgn7 | -0.345 |
| atp5f1d | 0.438 | anp32e | -0.342 |
| CABZ01072309.1 | 0.437 | lig1 | -0.342 |
| cox4i1 | 0.437 | top1l | -0.340 |
| cox8a | 0.436 | cirbpb | -0.338 |
| neb | 0.434 | hnrnpabb | -0.338 |