"ubiquinol-cytochrome c reductase, complex III subunit X"
ZFIN 
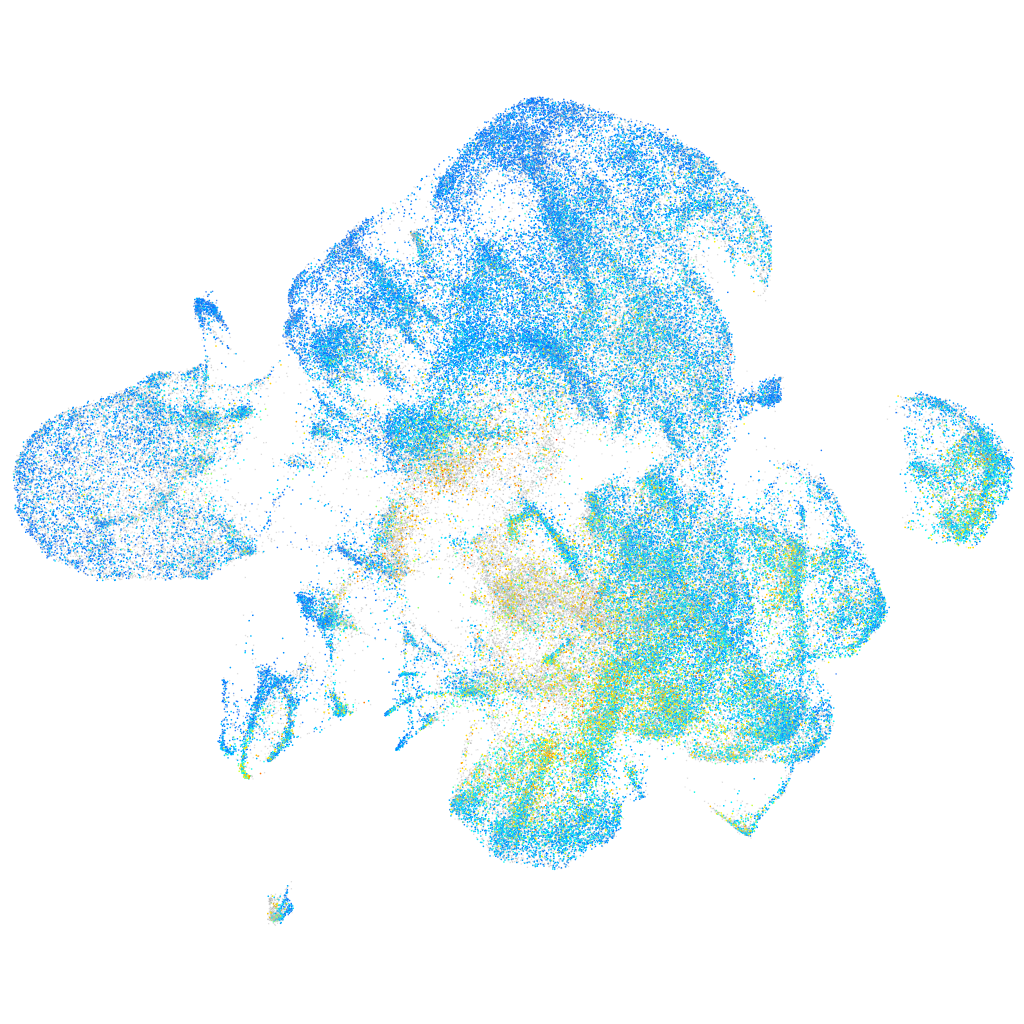
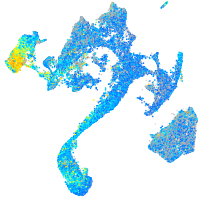
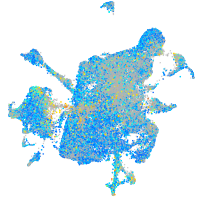
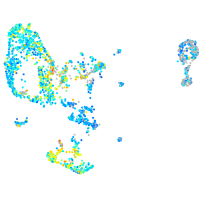
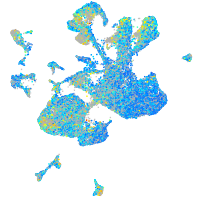
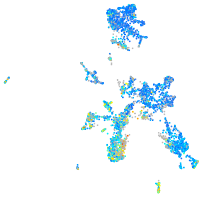
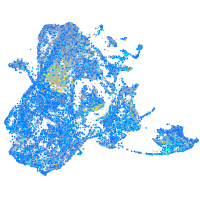
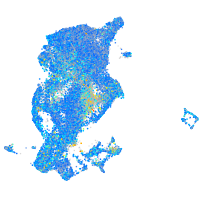
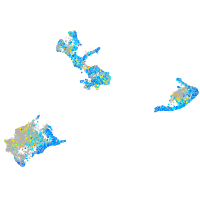
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5mc1 | 0.228 | hmgb2a | -0.111 |
| gapdhs | 0.216 | hspb1 | -0.104 |
| ppiab | 0.212 | si:ch211-152c2.3 | -0.100 |
| atp6v1g1 | 0.205 | stm | -0.094 |
| tpi1b | 0.203 | apoeb | -0.092 |
| atp6v1e1b | 0.198 | akap12b | -0.091 |
| ywhag2 | 0.197 | hmga1a | -0.089 |
| gng3 | 0.195 | NC-002333.4 | -0.088 |
| sncb | 0.190 | apoc1 | -0.088 |
| cox7a2a | 0.189 | aldob | -0.085 |
| ldhba | 0.187 | crabp2b | -0.085 |
| tuba1c | 0.186 | id1 | -0.081 |
| calm2a | 0.186 | apela | -0.081 |
| zgc:65894 | 0.185 | nr6a1a | -0.080 |
| calm1a | 0.185 | pou5f3 | -0.076 |
| stmn2a | 0.182 | zgc:110425 | -0.075 |
| ckbb | 0.182 | ucp2 | -0.074 |
| atp5l | 0.180 | polr3gla | -0.074 |
| cox8a | 0.177 | cx43.4 | -0.073 |
| mllt11 | 0.176 | si:dkey-66i24.9 | -0.071 |
| calm3b | 0.175 | s100a1 | -0.070 |
| tmsb2 | 0.174 | alcamb | -0.070 |
| eno1a | 0.171 | rsl1d1 | -0.069 |
| zgc:153426 | 0.170 | mki67 | -0.068 |
| atp5f1b | 0.170 | arf1 | -0.068 |
| atp5pf | 0.170 | lig1 | -0.068 |
| ndufa4 | 0.169 | sfrp1a | -0.068 |
| vamp2 | 0.169 | msna | -0.068 |
| stxbp1a | 0.169 | eif4ebp3l | -0.067 |
| COX5B | 0.168 | vox | -0.067 |
| cnrip1a | 0.168 | COX7A2 | -0.067 |
| ywhah | 0.167 | si:dkey-42i9.4 | -0.067 |
| cox4i1 | 0.166 | sb:cb81 | -0.067 |
| atp5meb | 0.165 | shisa2a | -0.066 |
| atp5if1b | 0.165 | pkdccb | -0.066 |