"UDP glucuronosyltransferase 5 family, polypeptide B1"
ZFIN 
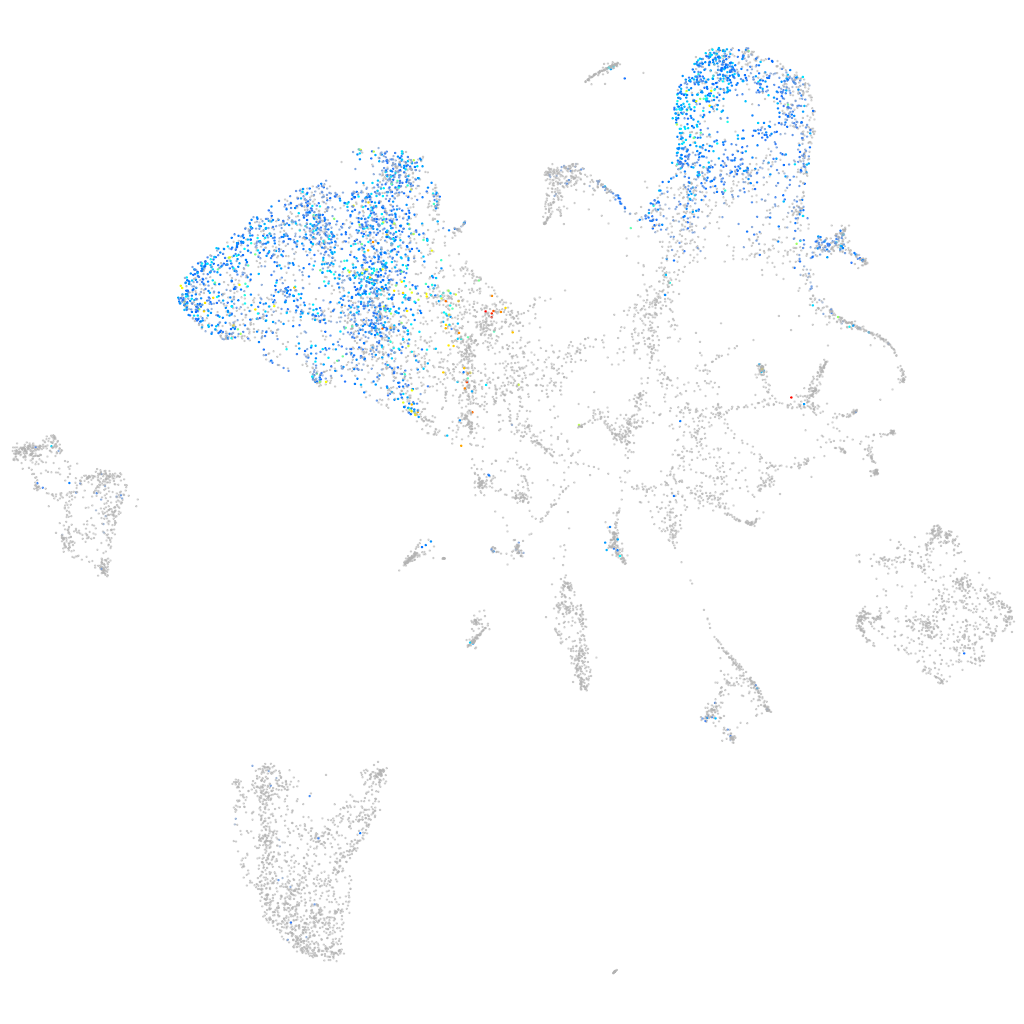
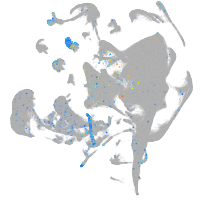
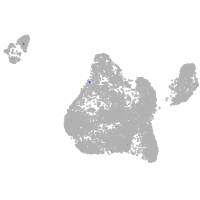
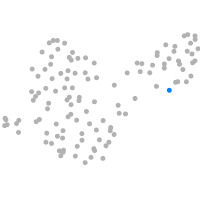
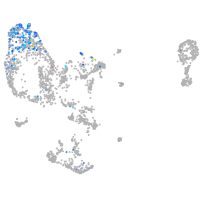
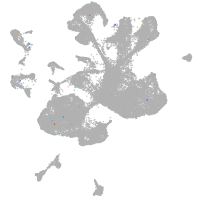
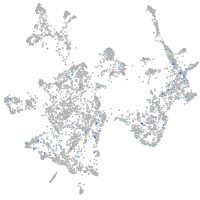
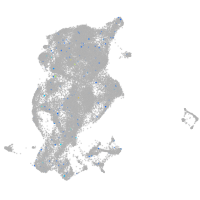
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ugt5b4 | 0.509 | si:ch211-222l21.1 | -0.268 |
| apoa4b.1 | 0.488 | hmgn6 | -0.246 |
| dhrs9 | 0.485 | slc38a5b | -0.246 |
| cyp2n13 | 0.485 | h3f3d | -0.243 |
| rdh1 | 0.473 | epcam | -0.241 |
| apoc2 | 0.472 | idh2 | -0.241 |
| scp2a | 0.471 | si:ch73-1a9.3 | -0.237 |
| ugt1a7 | 0.471 | h2afvb | -0.234 |
| gpx4a | 0.470 | hmgn2 | -0.232 |
| mgst1.2 | 0.460 | hmgb3a | -0.232 |
| apobb.1 | 0.460 | hmgb2a | -0.232 |
| glud1b | 0.458 | cirbpa | -0.232 |
| cyp4v8 | 0.457 | cirbpb | -0.231 |
| ckba | 0.449 | ptmab | -0.231 |
| etnppl | 0.440 | rtn1a | -0.226 |
| slc37a4a | 0.437 | eef1a1l1 | -0.226 |
| srd5a2a | 0.437 | ubc | -0.225 |
| cyp8b1 | 0.434 | khdrbs1a | -0.225 |
| slco1d1 | 0.432 | tuba8l4 | -0.222 |
| cyp2ad2 | 0.427 | syncrip | -0.215 |
| abat | 0.426 | ywhaz | -0.215 |
| sdr16c5b | 0.425 | hnrnpaba | -0.213 |
| agmo | 0.424 | glulb | -0.213 |
| afp4 | 0.424 | nucks1a | -0.212 |
| sult2st2 | 0.423 | hmgb1b | -0.210 |
| gstt1a | 0.422 | si:ch73-281n10.2 | -0.206 |
| gapdh | 0.421 | jpt1b | -0.206 |
| fbp1b | 0.419 | hmgb2b | -0.205 |
| gcshb | 0.418 | si:ch211-195b11.3 | -0.205 |
| pgam1a | 0.418 | hmgb1a | -0.205 |
| suclg1 | 0.416 | cldnb | -0.202 |
| haao | 0.415 | aldoaa | -0.202 |
| tdo2a | 0.415 | anp32b | -0.201 |
| mdh1aa | 0.414 | hmga1a | -0.199 |
| cat | 0.414 | marcksl1b | -0.199 |