ubiquitin domain containing 1b
ZFIN 
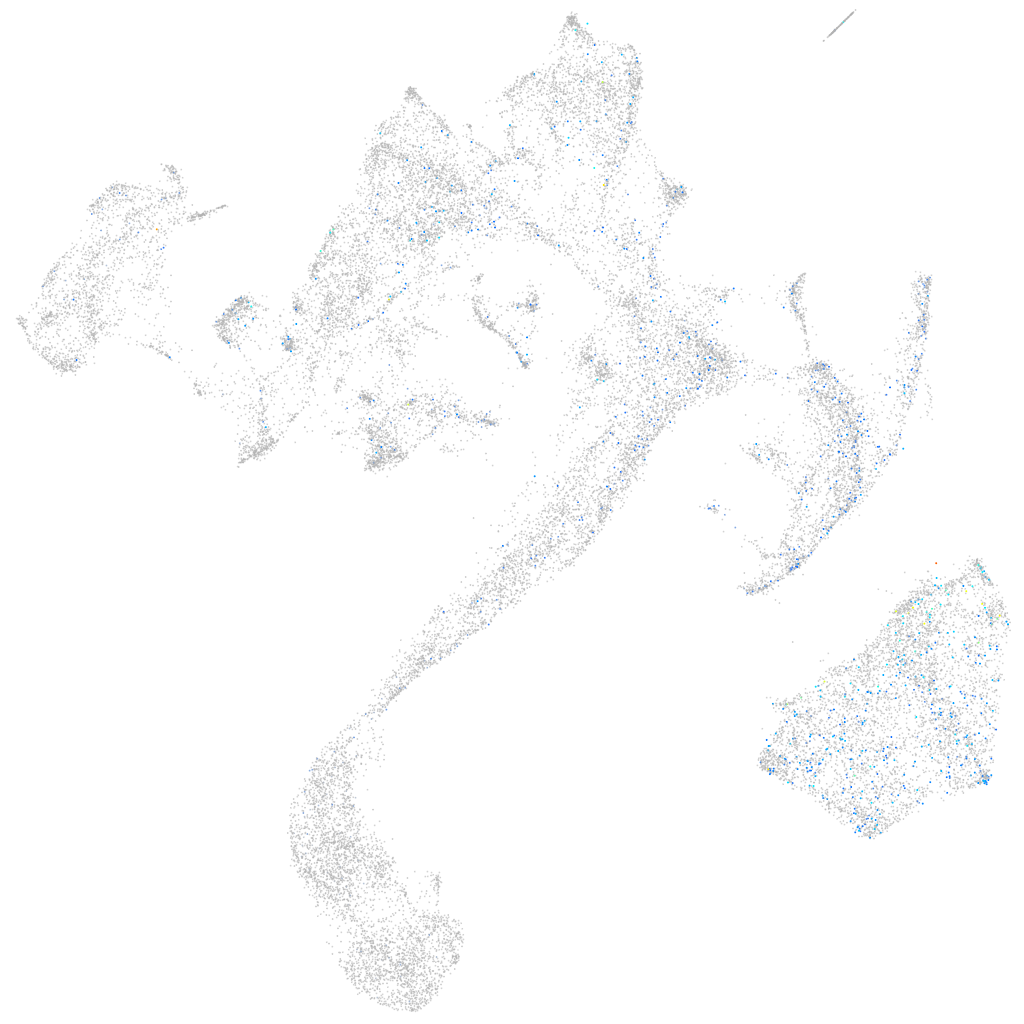
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpub | 0.128 | rpl37 | -0.118 |
| hnrnpa1b | 0.126 | zgc:114188 | -0.109 |
| nucks1a | 0.122 | rps10 | -0.108 |
| snrnp70 | 0.121 | rps17 | -0.105 |
| NC-002333.4 | 0.119 | actc1b | -0.097 |
| cdx4 | 0.119 | ak1 | -0.086 |
| anp32e | 0.119 | atp2a1 | -0.085 |
| marcksb | 0.119 | fabp3 | -0.085 |
| ppm1g | 0.118 | ckmb | -0.085 |
| hnrnpa1a | 0.118 | ckma | -0.084 |
| ilf3b | 0.118 | vdac2 | -0.082 |
| seta | 0.117 | atp5meb | -0.082 |
| rbm4.3 | 0.116 | pabpc4 | -0.082 |
| ppig | 0.115 | mt-nd1 | -0.081 |
| ncl | 0.115 | tmem38a | -0.080 |
| hnrnpm | 0.115 | aldoab | -0.080 |
| acin1a | 0.113 | atp5f1b | -0.080 |
| stm | 0.113 | ttn.2 | -0.079 |
| cx43.4 | 0.112 | mylpfa | -0.079 |
| safb | 0.112 | eef1da | -0.078 |
| zgc:110425 | 0.111 | tnnc2 | -0.078 |
| arf1 | 0.111 | gamt | -0.078 |
| syncrip | 0.111 | pvalb2 | -0.076 |
| prpf40a | 0.110 | atp5if1b | -0.075 |
| pnn | 0.110 | suclg1 | -0.075 |
| chd4b | 0.110 | acta1b | -0.075 |
| hmga1a | 0.110 | mylz3 | -0.075 |
| top1l | 0.110 | vdac3 | -0.075 |
| srsf1a | 0.109 | neb | -0.075 |
| fbl | 0.109 | tpma | -0.075 |
| thrap3b | 0.108 | pvalb1 | -0.075 |
| cirbpa | 0.108 | eno3 | -0.075 |
| ppp2cb | 0.108 | nme2b.2 | -0.074 |
| hnrnpd | 0.108 | eef1b2 | -0.074 |
| anp32a | 0.107 | ndufa3 | -0.074 |