tumor suppressor candidate 3
ZFIN 
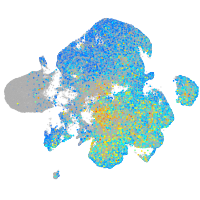
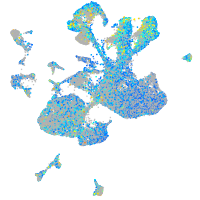
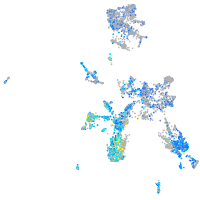
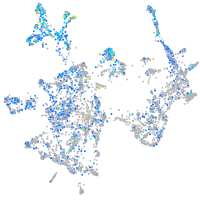
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.343 | gapdh | -0.210 |
| pcsk1nl | 0.328 | gamt | -0.187 |
| rnasekb | 0.300 | aldob | -0.183 |
| neurod1 | 0.294 | fbp1b | -0.181 |
| gapdhs | 0.293 | ahcy | -0.179 |
| syt1a | 0.289 | gatm | -0.174 |
| scgn | 0.279 | apoa4b.1 | -0.172 |
| rims2a | 0.276 | apoc2 | -0.168 |
| gpc1a | 0.274 | gstt1a | -0.165 |
| LOC100537384 | 0.271 | scp2a | -0.162 |
| mir7a-1 | 0.270 | mat1a | -0.156 |
| si:dkey-153k10.9 | 0.268 | eno3 | -0.156 |
| gnao1a | 0.265 | afp4 | -0.155 |
| lysmd2 | 0.263 | apoa1b | -0.155 |
| atp6v0cb | 0.262 | glud1b | -0.153 |
| jpt1b | 0.260 | pnp4b | -0.147 |
| pcsk2 | 0.259 | agxtb | -0.146 |
| atpv0e2 | 0.257 | suclg2 | -0.146 |
| pcsk1 | 0.255 | cx32.3 | -0.146 |
| pax6b | 0.255 | sult2st2 | -0.145 |
| kcnj11 | 0.253 | apobb.1 | -0.145 |
| egr4 | 0.252 | apoc1 | -0.144 |
| vamp2 | 0.251 | ugt1a7 | -0.144 |
| vat1 | 0.250 | gpx4a | -0.144 |
| gdi1 | 0.249 | rdh1 | -0.142 |
| zgc:101731 | 0.248 | tdo2a | -0.142 |
| rprmb | 0.247 | mgst1.2 | -0.140 |
| id4 | 0.246 | cdo1 | -0.140 |
| cpe | 0.246 | msrb2 | -0.140 |
| tspan7b | 0.245 | aldh6a1 | -0.140 |
| aldocb | 0.244 | bhmt | -0.139 |
| jagn1a | 0.243 | gnmt | -0.138 |
| rasd4 | 0.242 | gcshb | -0.138 |
| insm1a | 0.241 | cox7a1 | -0.138 |
| scg2a | 0.238 | agxta | -0.137 |