"tubulin, alpha 8 like 3"
ZFIN 
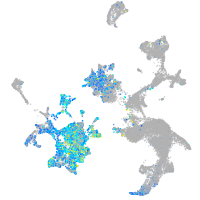
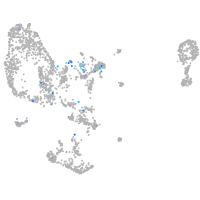
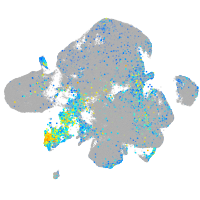
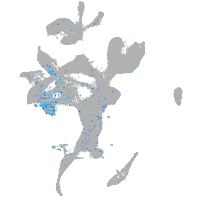
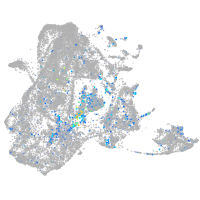
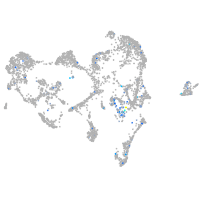
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| six2a | 0.198 | aldob | -0.077 |
| LOC101885084 | 0.182 | eno3 | -0.072 |
| marcksl1a | 0.180 | aldh6a1 | -0.071 |
| LOC110440219 | 0.175 | abat | -0.069 |
| defbl1 | 0.175 | haao | -0.067 |
| qkib | 0.174 | mt-nd2 | -0.067 |
| sparc | 0.169 | gstr | -0.066 |
| si:ch211-243a20.3 | 0.163 | pklr | -0.066 |
| pnp4a | 0.158 | slc37a4a | -0.066 |
| qki2 | 0.157 | aqp12 | -0.065 |
| si:ch1073-268j14.1 | 0.154 | cat | -0.064 |
| apoda.1 | 0.150 | agxtb | -0.064 |
| six1b | 0.149 | lipf | -0.064 |
| LOC110439013 | 0.144 | g6pca.2 | -0.063 |
| phox2bb | 0.142 | suclg2 | -0.063 |
| tlx2 | 0.141 | glud1b | -0.063 |
| LOC103909099 | 0.140 | pnp4b | -0.063 |
| zeb2a | 0.138 | pgm1 | -0.063 |
| ccdc80 | 0.135 | mt-nd5 | -0.063 |
| elavl3 | 0.135 | sb:cb1058 | -0.062 |
| mcamb | 0.135 | agxta | -0.062 |
| timd4 | 0.135 | cox7a1 | -0.062 |
| skor1a | 0.135 | nipsnap3a | -0.062 |
| scrt2 | 0.133 | mt-nd4 | -0.062 |
| si:ch211-218m3.11 | 0.133 | gpd1b | -0.062 |
| nrcamb | 0.131 | dap | -0.062 |
| rbfox1 | 0.130 | pck2 | -0.061 |
| cyr61 | 0.127 | rltgr | -0.061 |
| edn1 | 0.126 | gapdh | -0.061 |
| sim2.1 | 0.126 | isoc2 | -0.061 |
| unm-sa821 | 0.126 | scp2a | -0.060 |
| hyal4 | 0.126 | hdlbpa | -0.060 |
| XLOC-019062 | 0.125 | as3mt | -0.060 |
| zmat4b | 0.125 | ahcy | -0.060 |
| syngr2b | 0.125 | gstt1a | -0.060 |