tetraspanin 9b
ZFIN 
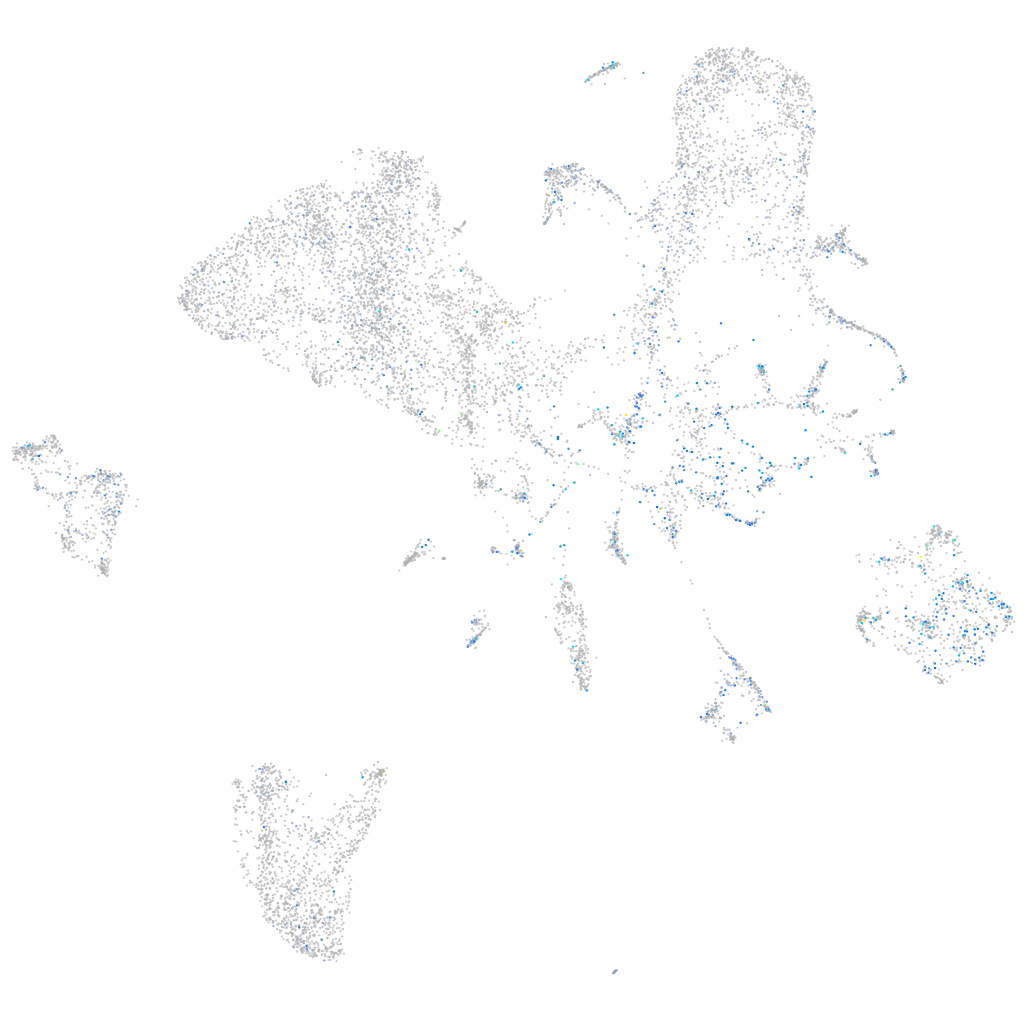
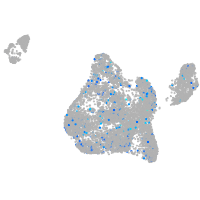
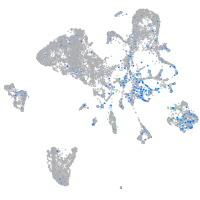
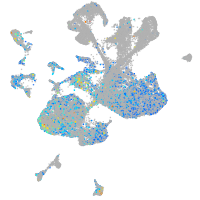
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgn6 | 0.177 | gamt | -0.160 |
| hnrnpaba | 0.176 | gapdh | -0.157 |
| khdrbs1a | 0.174 | ahcy | -0.150 |
| h3f3d | 0.172 | gatm | -0.148 |
| si:ch211-222l21.1 | 0.170 | bhmt | -0.140 |
| syncrip | 0.164 | mat1a | -0.137 |
| nucks1a | 0.162 | fbp1b | -0.132 |
| hdac1 | 0.162 | apoa1b | -0.131 |
| ubc | 0.161 | apoa4b.1 | -0.129 |
| si:ch73-1a9.3 | 0.160 | apoc2 | -0.128 |
| ctnnb1 | 0.160 | aldob | -0.126 |
| si:dkey-42i9.4 | 0.159 | nupr1b | -0.125 |
| h2afvb | 0.159 | afp4 | -0.123 |
| acin1a | 0.158 | gpx4a | -0.120 |
| hmga1a | 0.156 | scp2a | -0.120 |
| hmgb3a | 0.156 | suclg1 | -0.120 |
| marcksl1b | 0.155 | pnp4b | -0.118 |
| cirbpb | 0.155 | agxtb | -0.117 |
| hp1bp3 | 0.154 | aldh7a1 | -0.116 |
| jpt1b | 0.154 | glud1b | -0.116 |
| akap12b | 0.154 | eno3 | -0.115 |
| smc1al | 0.153 | apoa2 | -0.115 |
| hmgb2a | 0.153 | apobb.1 | -0.114 |
| hmgn7 | 0.152 | gstt1a | -0.112 |
| tspan7 | 0.151 | aqp12 | -0.111 |
| cbx3a | 0.151 | cx32.3 | -0.108 |
| si:ch73-281n10.2 | 0.151 | grhprb | -0.107 |
| marcksb | 0.151 | dap | -0.106 |
| seta | 0.151 | suclg2 | -0.106 |
| cirbpa | 0.150 | pgm1 | -0.106 |
| stm | 0.149 | rbp4 | -0.105 |
| cdh6 | 0.149 | nipsnap3a | -0.104 |
| hnrnpa0b | 0.149 | agxta | -0.104 |
| tpm4a | 0.149 | fabp10a | -0.104 |
| id1 | 0.149 | mdh1aa | -0.103 |