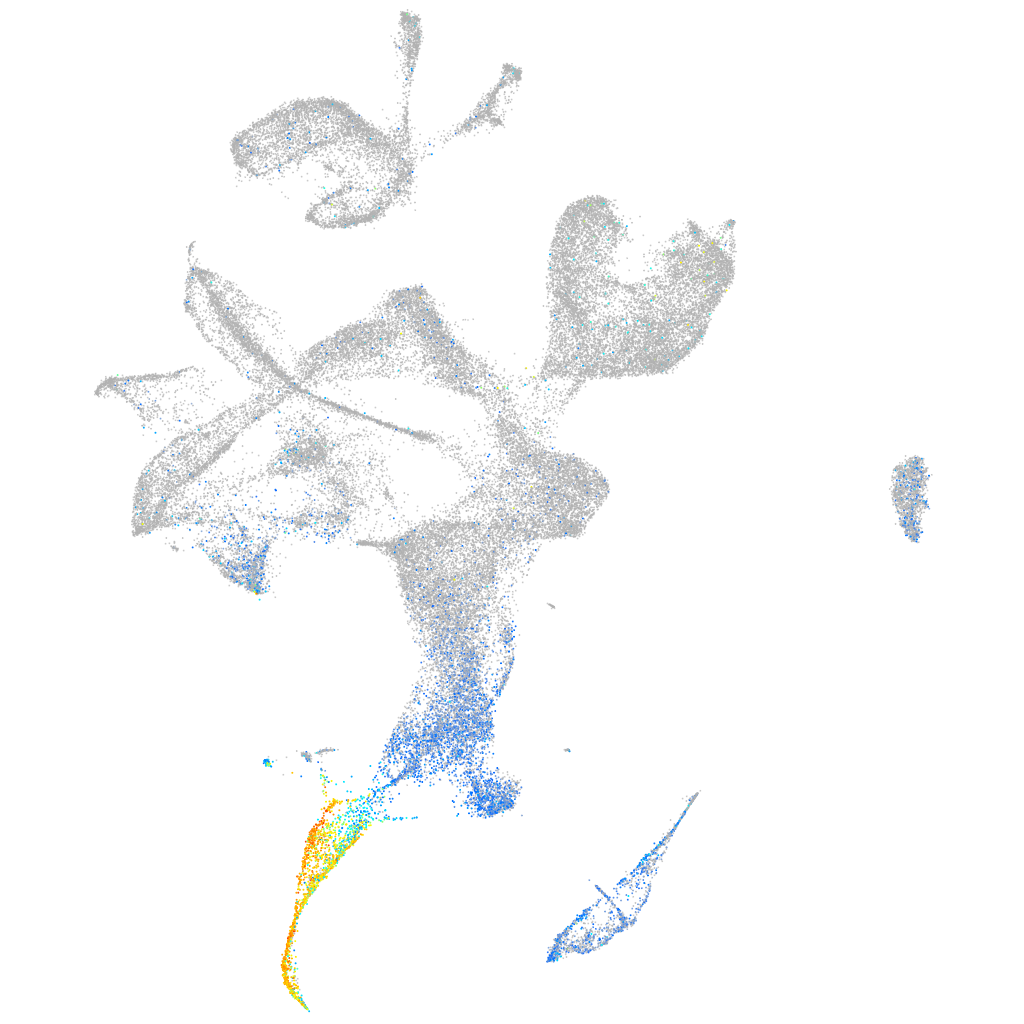tetraspanin 36
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dct | 0.849 | ptmab | -0.424 |
| pmela | 0.831 | si:ch73-1a9.3 | -0.337 |
| tyrp1b | 0.827 | hmgn6 | -0.278 |
| slc45a2 | 0.798 | ptmaa | -0.272 |
| pmelb | 0.795 | tuba1c | -0.254 |
| gpr143 | 0.779 | gpm6aa | -0.244 |
| tyrp1a | 0.767 | hmgb1a | -0.219 |
| tspan10 | 0.742 | marcksl1b | -0.213 |
| oca2 | 0.742 | si:ch211-137a8.4 | -0.195 |
| bace2 | 0.720 | tuba1a | -0.188 |
| rab38 | 0.707 | fabp7a | -0.187 |
| fabp11b | 0.679 | cadm3 | -0.181 |
| rgrb | 0.676 | epb41a | -0.178 |
| pttg1ipb | 0.672 | h2afva | -0.177 |
| cracr2ab | 0.666 | nova2 | -0.177 |
| slc24a5 | 0.655 | anp32e | -0.175 |
| qdpra | 0.653 | ckbb | -0.173 |
| gstp1 | 0.646 | snap25b | -0.172 |
| tyr | 0.638 | hmgn2 | -0.170 |
| msnb | 0.630 | hmgb3a | -0.168 |
| agtrap | 0.625 | si:ch1073-429i10.3.1 | -0.166 |
| zgc:110591 | 0.620 | gapdhs | -0.162 |
| col4a5 | 0.609 | foxg1b | -0.161 |
| rab27a | 0.607 | rtn1a | -0.159 |
| sparc | 0.606 | rorb | -0.159 |
| stra6 | 0.605 | hmgb1b | -0.159 |
| LOC103910009 | 0.604 | stmn1b | -0.159 |
| tm6sf2 | 0.604 | mdkb | -0.157 |
| mitfa | 0.598 | crx | -0.153 |
| fgfbp1a | 0.597 | marcksl1a | -0.152 |
| trpm1b | 0.592 | h3f3d | -0.149 |
| mb | 0.589 | neurod4 | -0.149 |
| col4a6 | 0.584 | hnrnpaba | -0.148 |
| sytl2a | 0.581 | h3f3c | -0.148 |
| triobpa | 0.576 | atp1b2a | -0.147 |