teashirt zinc finger homeobox 3b
ZFIN 
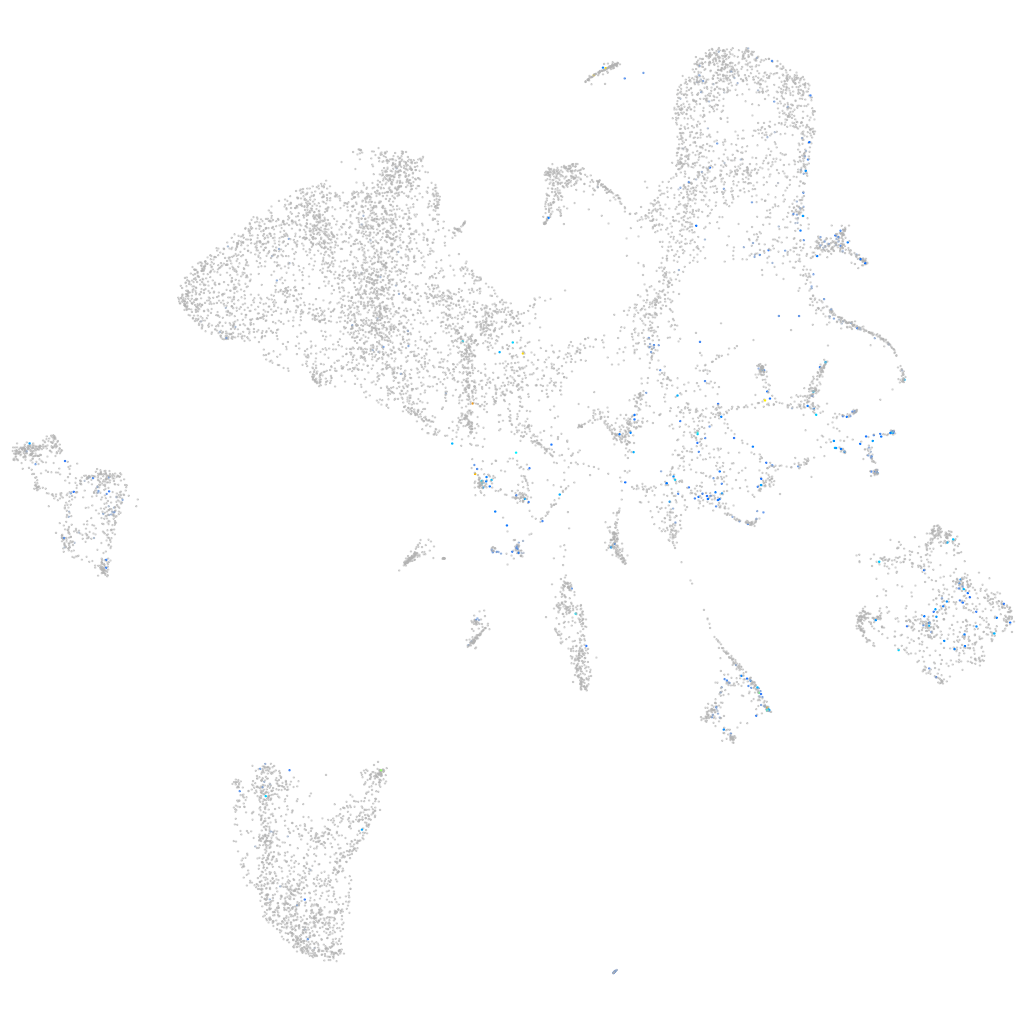
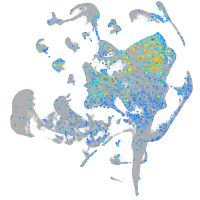
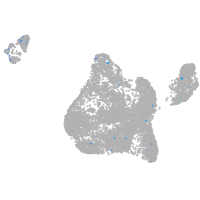
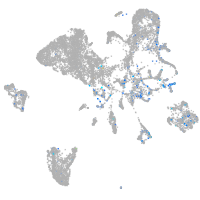
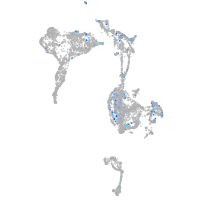
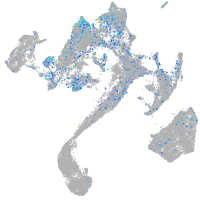
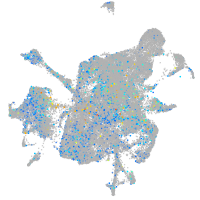
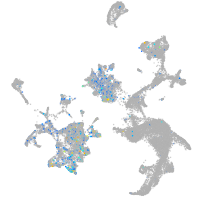
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| XLOC-043912 | 0.175 | gamt | -0.119 |
| LOC110440001 | 0.172 | gapdh | -0.116 |
| zbbx | 0.158 | ahcy | -0.115 |
| selenoo2 | 0.155 | gatm | -0.114 |
| LOC101886524 | 0.149 | aldob | -0.105 |
| lrrc18b | 0.144 | aldh7a1 | -0.103 |
| chrna2b | 0.141 | mat1a | -0.103 |
| dpp6a | 0.134 | bhmt | -0.102 |
| jpt1b | 0.132 | apoa1b | -0.102 |
| zgc:92818 | 0.131 | gpx4a | -0.101 |
| kcnd1 | 0.127 | glud1b | -0.101 |
| si:dkeyp-75h12.5 | 0.126 | pnp4b | -0.100 |
| cacng6b | 0.126 | apoc2 | -0.098 |
| si:dkey-47k16.4 | 0.125 | apoa4b.1 | -0.097 |
| cplx2l | 0.125 | nipsnap3a | -0.095 |
| sim2 | 0.124 | cx32.3 | -0.095 |
| atp8b4 | 0.121 | fbp1b | -0.094 |
| necap1 | 0.121 | aqp12 | -0.094 |
| col4a4 | 0.120 | grhprb | -0.093 |
| neurod1 | 0.120 | sod1 | -0.093 |
| sema6ba | 0.118 | eno3 | -0.093 |
| adgrl3.1 | 0.118 | agxtb | -0.092 |
| vat1 | 0.118 | nupr1b | -0.092 |
| CABZ01055365.1 | 0.118 | suclg1 | -0.092 |
| LOC101885084 | 0.117 | fgb | -0.091 |
| marcksl1b | 0.116 | serpina1l | -0.091 |
| atrx | 0.115 | afp4 | -0.089 |
| fstl1b | 0.115 | fga | -0.089 |
| tshz3a | 0.115 | fabp10a | -0.089 |
| hmgn6 | 0.114 | pgm1 | -0.088 |
| ksr2 | 0.113 | zgc:123103 | -0.088 |
| glra1 | 0.112 | serpina1 | -0.088 |
| hmgb1a | 0.112 | ces2 | -0.088 |
| efnb2a | 0.112 | apobb.1 | -0.087 |
| pnp4a | 0.111 | scp2a | -0.087 |